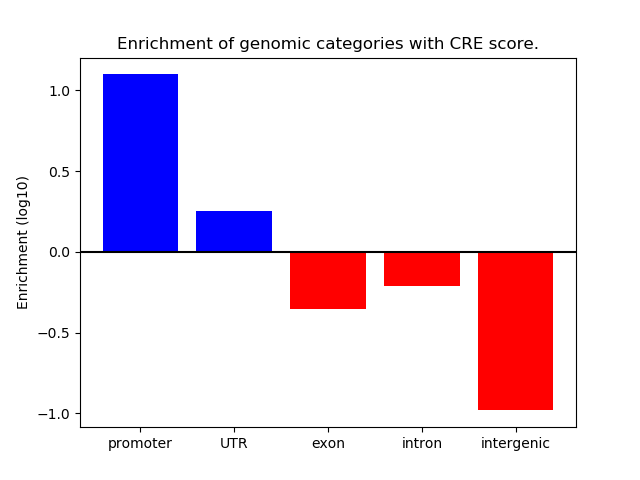
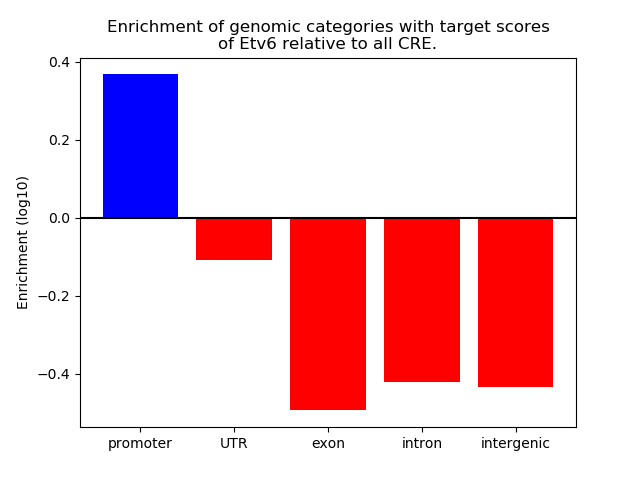
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
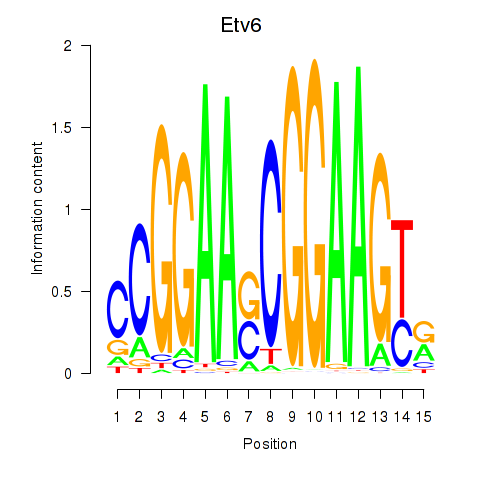
Results for Etv6
Z-value: 0.50
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.10 | ets variant 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_134211159_134211530 | Etv6 | 22256 | 0.149474 | -0.40 | 1.8e-03 | Click! |
| chr6_134035013_134035747 | Etv6 | 320 | 0.899920 | 0.34 | 8.6e-03 | Click! |
| chr6_134042024_134042311 | Etv6 | 6198 | 0.206162 | -0.30 | 2.0e-02 | Click! |
| chr6_134035835_134036398 | Etv6 | 147 | 0.961428 | 0.29 | 2.7e-02 | Click! |
| chr6_134037752_134037903 | Etv6 | 1858 | 0.355570 | -0.28 | 3.3e-02 | Click! |
Activity of the Etv6 motif across conditions
Conditions sorted by the z-value of the Etv6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_78180060_78180244 | 1.09 |
Tlcd1 |
TLC domain containing 1 |
879 |
0.2 |
| chr5_125361044_125361231 | 1.08 |
Gm40323 |
predicted gene, 40323 |
38 |
0.96 |
| chr17_83808858_83809009 | 1.03 |
Mta3 |
metastasis associated 3 |
13511 |
0.18 |
| chr2_148680576_148680950 | 1.00 |
Gzf1 |
GDNF-inducible zinc finger protein 1 |
260 |
0.9 |
| chr11_5485603_5486347 | 0.98 |
Gm11962 |
predicted gene 11962 |
9471 |
0.14 |
| chr6_125009489_125010173 | 0.98 |
Zfp384 |
zinc finger protein 384 |
26 |
0.94 |
| chr7_45016836_45018032 | 0.91 |
Rras |
related RAS viral (r-ras) oncogene |
527 |
0.43 |
| chr11_95011480_95011953 | 0.88 |
Samd14 |
sterile alpha motif domain containing 14 |
1435 |
0.27 |
| chr8_122556522_122557920 | 0.88 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
5892 |
0.1 |
| chr19_53257195_53257790 | 0.84 |
1700001K23Rik |
RIKEN cDNA 1700001K23 gene |
2272 |
0.25 |
| chr1_45794912_45795350 | 0.78 |
Wdr75 |
WD repeat domain 75 |
35 |
0.97 |
| chr7_141209842_141210144 | 0.78 |
Lmntd2 |
lamin tail domain containing 2 |
4084 |
0.08 |
| chr15_80096554_80097109 | 0.76 |
Syngr1 |
synaptogyrin 1 |
1039 |
0.33 |
| chr15_83030057_83030676 | 0.75 |
Nfam1 |
Nfat activating molecule with ITAM motif 1 |
416 |
0.79 |
| chr7_28796055_28797204 | 0.74 |
Rinl |
Ras and Rab interactor-like |
309 |
0.76 |
| chr15_85811807_85811994 | 0.73 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
203 |
0.91 |
| chr1_180225593_180226807 | 0.71 |
Psen2 |
presenilin 2 |
3368 |
0.18 |
| chr14_55671318_55672155 | 0.71 |
Nedd8 |
neural precursor cell expressed, developmentally down-regulated gene 8 |
137 |
0.55 |
| chr7_105743752_105744690 | 0.70 |
Taf10 |
TATA-box binding protein associated factor 10 |
121 |
0.91 |
| chr4_115850002_115850153 | 0.70 |
Mknk1 |
MAP kinase-interacting serine/threonine kinase 1 |
6955 |
0.13 |
| chr6_87670319_87670941 | 0.69 |
E230015B07Rik |
RIKEN cDNA E230015B07 gene |
1353 |
0.24 |
| chr11_97511052_97512791 | 0.68 |
Gm11611 |
predicted gene 11611 |
9974 |
0.12 |
| chr11_77792865_77793937 | 0.67 |
Gm10277 |
predicted gene 10277 |
5654 |
0.16 |
| chr15_78877218_78878328 | 0.67 |
Gga1 |
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
222 |
0.83 |
| chr16_56037323_56037733 | 0.64 |
Trmt10c |
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
291 |
0.8 |
| chr11_114668556_114669119 | 0.64 |
Rpl38 |
ribosomal protein L38 |
211 |
0.92 |
| chr15_97379421_97380753 | 0.63 |
Pced1b |
PC-esterase domain containing 1B |
18870 |
0.24 |
| chr15_78876971_78877162 | 0.61 |
Gga1 |
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
124 |
0.9 |
| chr4_98383218_98383435 | 0.61 |
Tm2d1 |
TM2 domain containing 1 |
20 |
0.67 |
| chr2_126932377_126933507 | 0.60 |
Sppl2a |
signal peptide peptidase like 2A |
293 |
0.9 |
| chr9_13245250_13245857 | 0.58 |
Jrkl |
Jrk-like |
276 |
0.86 |
| chr1_193172818_193173434 | 0.58 |
A130010J15Rik |
RIKEN cDNA A130010J15 gene |
342 |
0.79 |
| chr7_4149072_4149804 | 0.58 |
Leng9 |
leukocyte receptor cluster (LRC) member 9 |
936 |
0.34 |
| chr5_147860597_147861358 | 0.57 |
Pomp |
proteasome maturation protein |
324 |
0.87 |
| chr8_94182622_94183855 | 0.57 |
Gm39228 |
predicted gene, 39228 |
51 |
0.95 |
| chr9_20581003_20581881 | 0.56 |
Zfp846 |
zinc finger protein 846 |
83 |
0.95 |
| chr11_96915727_96916437 | 0.56 |
Cdk5rap3 |
CDK5 regulatory subunit associated protein 3 |
25 |
0.69 |
| chr5_123150083_123151176 | 0.55 |
Setd1b |
SET domain containing 1B |
7672 |
0.08 |
| chr17_35135244_35136480 | 0.55 |
Bag6 |
BCL2-associated athanogene 6 |
118 |
0.84 |
| chr12_13180713_13181236 | 0.54 |
Gm24208 |
predicted gene, 24208 |
17211 |
0.16 |
| chr12_37241627_37242172 | 0.54 |
Agmo |
alkylglycerol monooxygenase |
39 |
0.99 |
| chr7_127471033_127472292 | 0.53 |
Prr14 |
proline rich 14 |
172 |
0.84 |
| chr4_82858859_82859877 | 0.52 |
Zdhhc21 |
zinc finger, DHHC domain containing 21 |
186 |
0.95 |
| chr11_120098951_120099839 | 0.52 |
Ndufaf8 |
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
296 |
0.68 |
| chr8_84977210_84978736 | 0.52 |
Junb |
jun B proto-oncogene |
745 |
0.32 |
| chr12_3235357_3235508 | 0.52 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
72 |
0.97 |
| chr6_120364033_120365030 | 0.52 |
Kdm5a |
lysine (K)-specific demethylase 5A |
73 |
0.62 |
| chr2_132686926_132687263 | 0.52 |
Shld1 |
shieldin complex subunit 1 |
163 |
0.92 |
| chr2_181365439_181366324 | 0.50 |
Zgpat |
zinc finger, CCCH-type with G patch domain |
429 |
0.44 |
| chr18_68229517_68229916 | 0.48 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
1649 |
0.37 |
| chr6_56797213_56797748 | 0.48 |
Gm10209 |
predicted gene 10209 |
111 |
0.63 |
| chr11_58104070_58104840 | 0.48 |
Cnot8 |
CCR4-NOT transcription complex, subunit 8 |
89 |
0.95 |
| chr9_108490802_108491599 | 0.48 |
Usp19 |
ubiquitin specific peptidase 19 |
344 |
0.67 |
| chr11_48816327_48816915 | 0.48 |
Trim41 |
tripartite motif-containing 41 |
332 |
0.76 |
| chr17_86300838_86301701 | 0.48 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
14091 |
0.27 |
| chr5_146948357_146948548 | 0.47 |
Gtf3a |
general transcription factor III A |
205 |
0.93 |
| chr9_19622148_19622587 | 0.47 |
Zfp317 |
zinc finger protein 317 |
63 |
0.96 |
| chr17_21733400_21733702 | 0.47 |
Zfp229 |
zinc finger protein 229 |
129 |
0.94 |
| chr17_29263369_29264286 | 0.47 |
Ppil1 |
peptidylprolyl isomerase (cyclophilin)-like 1 |
111 |
0.58 |
| chr11_62005417_62006894 | 0.46 |
Specc1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
805 |
0.61 |
| chr1_131136063_131136570 | 0.46 |
Dyrk3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
1929 |
0.25 |
| chr7_13033778_13034709 | 0.46 |
Chmp2a |
charged multivesicular body protein 2A |
401 |
0.66 |
| chr10_20952007_20952498 | 0.45 |
Ahi1 |
Abelson helper integration site 1 |
295 |
0.92 |
| chr8_110217965_110218603 | 0.45 |
Cmtr2 |
cap methyltransferase 2 |
295 |
0.9 |
| chr15_73156643_73156955 | 0.44 |
Ago2 |
argonaute RISC catalytic subunit 2 |
21048 |
0.18 |
| chr8_70139467_70139991 | 0.44 |
Borcs8 |
BLOC-1 related complex subunit 8 |
6 |
0.47 |
| chr4_133277237_133277725 | 0.43 |
Tmem222 |
transmembrane protein 222 |
137 |
0.93 |
| chr10_62327314_62328305 | 0.43 |
Hk1 |
hexokinase 1 |
42 |
0.97 |
| chr2_73058170_73058361 | 0.42 |
Gm13665 |
predicted gene 13665 |
58483 |
0.09 |
| chrX_42067935_42068524 | 0.42 |
Xiap |
X-linked inhibitor of apoptosis |
169 |
0.96 |
| chr9_70421677_70422108 | 0.42 |
Ccnb2 |
cyclin B2 |
345 |
0.85 |
| chr2_12419525_12419707 | 0.42 |
Mindy3 |
MINDY lysine 48 deubiquitinase 3 |
146 |
0.94 |
| chr4_125066307_125067492 | 0.41 |
Snip1 |
Smad nuclear interacting protein 1 |
197 |
0.86 |
| chr1_58505065_58505587 | 0.41 |
Gm15834 |
predicted gene 15834 |
171 |
0.55 |
| chr1_74600666_74601851 | 0.41 |
Rnf25 |
ring finger protein 25 |
87 |
0.63 |
| chr2_84714065_84715116 | 0.40 |
Zdhhc5 |
zinc finger, DHHC domain containing 5 |
96 |
0.92 |
| chr11_78031310_78031604 | 0.39 |
Dhrs13 |
dehydrogenase/reductase (SDR family) member 13 |
823 |
0.34 |
| chr9_46273030_46273181 | 0.39 |
Zpr1 |
ZPR1 zinc finger |
41 |
0.93 |
| chr3_89430003_89431210 | 0.39 |
Pygo2 |
pygopus 2 |
134 |
0.9 |
| chr2_27539910_27541089 | 0.39 |
Gm13421 |
predicted gene 13421 |
70 |
0.9 |
| chr17_33915601_33916103 | 0.39 |
Tapbp |
TAP binding protein |
47 |
0.48 |
| chr16_78560111_78560350 | 0.39 |
D16Ertd472e |
DNA segment, Chr 16, ERATO Doi 472, expressed |
14 |
0.56 |
| chr13_41097843_41097994 | 0.39 |
Gm48344 |
predicted gene, 48344 |
6488 |
0.13 |
| chr8_70509737_70510396 | 0.39 |
Uba52 |
ubiquitin A-52 residue ribosomal protein fusion product 1 |
121 |
0.9 |
| chr11_44528978_44529659 | 0.38 |
Rnf145 |
ring finger protein 145 |
9013 |
0.18 |
| chr11_73137063_73138275 | 0.38 |
Haspin |
histone H3 associated protein kinase |
625 |
0.6 |
| chr3_127889422_127889573 | 0.38 |
Fam241a |
family with sequence similarity 241, member A |
6791 |
0.13 |
| chr11_45853066_45853434 | 0.37 |
Clint1 |
clathrin interactor 1 |
1286 |
0.38 |
| chr11_119392018_119392570 | 0.37 |
Rnf213 |
ring finger protein 213 |
806 |
0.49 |
| chr16_76365731_76366199 | 0.37 |
Nrip1 |
nuclear receptor interacting protein 1 |
7072 |
0.21 |
| chr3_54811140_54811438 | 0.37 |
Gm10254 |
predicted gene 10254 |
39 |
0.97 |
| chr10_128408634_128409894 | 0.36 |
Nabp2 |
nucleic acid binding protein 2 |
84 |
0.9 |
| chr15_99474673_99475009 | 0.36 |
Bcdin3d |
BCDIN3 domain containing |
111 |
0.93 |
| chr1_156523916_156524227 | 0.36 |
Csnk2a3 |
casein kinase 2 alpha 3 |
59 |
0.97 |
| chr11_78245687_78246240 | 0.35 |
Supt6 |
SPT6, histone chaperone and transcription elongation factor |
24 |
0.72 |
| chr2_24386288_24387076 | 0.35 |
Psd4 |
pleckstrin and Sec7 domain containing 4 |
65 |
0.97 |
| chr12_105784740_105785770 | 0.35 |
Papola |
poly (A) polymerase alpha |
383 |
0.86 |
| chr3_157533826_157535237 | 0.35 |
Zranb2 |
zinc finger, RAN-binding domain containing 2 |
106 |
0.84 |
| chr2_132253346_132253893 | 0.35 |
AV099323 |
expressed sequence AV099323 |
262 |
0.53 |
| chr11_26591574_26591760 | 0.35 |
Vrk2 |
vaccinia related kinase 2 |
62 |
0.97 |
| chr19_45005673_45006396 | 0.35 |
Twnk |
twinkle mtDNA helicase |
371 |
0.45 |
| chr6_141488896_141489047 | 0.34 |
Slco1c1 |
solute carrier organic anion transporter family, member 1c1 |
35397 |
0.18 |
| chr14_55094784_55095911 | 0.34 |
Thtpa |
thiamine triphosphatase |
563 |
0.54 |
| chr2_75937843_75939119 | 0.34 |
Ttc30b |
tetratricopeptide repeat domain 30B |
104 |
0.96 |
| chr4_88878434_88878641 | 0.34 |
Ifne |
interferon epsilon |
1664 |
0.16 |
| chr19_3282559_3283552 | 0.33 |
Mrpl21 |
mitochondrial ribosomal protein L21 |
0 |
0.51 |
| chr11_69914045_69915024 | 0.33 |
Gps2 |
G protein pathway suppressor 2 |
311 |
0.67 |
| chr14_52279628_52280152 | 0.33 |
Tox4 |
TOX high mobility group box family member 4 |
158 |
0.69 |
| chr15_5185304_5185980 | 0.33 |
Gm15938 |
predicted gene 15938 |
81 |
0.49 |
| chr6_136793393_136794589 | 0.33 |
H4f16 |
H4 histone 16 |
10424 |
0.09 |
| chrX_134541627_134541952 | 0.32 |
Timm8a1 |
translocase of inner mitochondrial membrane 8A1 |
76 |
0.95 |
| chr4_31963905_31964773 | 0.32 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
76 |
0.98 |
| chr17_47010961_47011192 | 0.32 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
520 |
0.76 |
| chr5_22612995_22613241 | 0.32 |
Rpl17-ps5 |
ribosomal protein L17, pseudogene 5 |
64 |
0.97 |
| chr2_136890459_136890749 | 0.32 |
Mkks |
McKusick-Kaufman syndrome |
550 |
0.38 |
| chr7_39449138_39450316 | 0.31 |
Zfp939 |
zinc finger protein 939 |
209 |
0.75 |
| chr13_21362484_21362864 | 0.31 |
Zscan12 |
zinc finger and SCAN domain containing 12 |
146 |
0.9 |
| chr14_101639989_101640746 | 0.31 |
Commd6 |
COMM domain containing 6 |
81 |
0.97 |
| chr17_56256141_56256492 | 0.31 |
Fem1a |
fem 1 homolog a |
494 |
0.62 |
| chr3_150515944_150516241 | 0.31 |
Gm6439 |
predicted gene 6439 |
11 |
0.99 |
| chr12_76362328_76362736 | 0.30 |
Zbtb25 |
zinc finger and BTB domain containing 25 |
5392 |
0.11 |
| chr19_6910112_6910674 | 0.30 |
Trmt112 |
tRNA methyltransferase 11-2 |
229 |
0.52 |
| chr7_100492685_100494805 | 0.30 |
Ucp2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
50 |
0.95 |
| chr14_55064836_55065251 | 0.30 |
Zfhx2 |
zinc finger homeobox 2 |
7191 |
0.08 |
| chr6_125152493_125152726 | 0.30 |
Iffo1 |
intermediate filament family orphan 1 |
739 |
0.38 |
| chr19_46135570_46137235 | 0.30 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
887 |
0.45 |
| chr6_119481820_119482341 | 0.30 |
Fbxl14 |
F-box and leucine-rich repeat protein 14 |
2412 |
0.3 |
| chr7_16416809_16416960 | 0.29 |
Gm45510 |
predicted gene 45510 |
263 |
0.82 |
| chr1_134494702_134495438 | 0.29 |
Rabif |
RAB interacting factor |
422 |
0.72 |
| chr16_21333322_21333876 | 0.29 |
Magef1 |
melanoma antigen family F, 1 |
243 |
0.93 |
| chr7_25718309_25719059 | 0.29 |
Ccdc97 |
coiled-coil domain containing 97 |
307 |
0.8 |
| chr11_84867576_84868235 | 0.29 |
Ggnbp2 |
gametogenetin binding protein 2 |
2297 |
0.18 |
| chrX_50566542_50567054 | 0.29 |
Firre |
functional intergenic repeating RNA element |
1915 |
0.4 |
| chr15_59317788_59318646 | 0.28 |
Sqle |
squalene epoxidase |
3110 |
0.21 |
| chrX_142389571_142390415 | 0.28 |
Acsl4 |
acyl-CoA synthetase long-chain family member 4 |
351 |
0.84 |
| chr6_128424733_128425194 | 0.28 |
Itfg2 |
integrin alpha FG-GAP repeat containing 2 |
32 |
0.94 |
| chr3_95436992_95437143 | 0.28 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
2548 |
0.14 |
| chr7_45434237_45434647 | 0.28 |
Ruvbl2 |
RuvB-like protein 2 |
165 |
0.68 |
| chr12_108791850_108792666 | 0.27 |
Gm34220 |
predicted gene, 34220 |
485 |
0.52 |
| chr5_146692514_146692846 | 0.27 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
13942 |
0.17 |
| chr2_180041752_180042359 | 0.27 |
Psma7 |
proteasome subunit alpha 7 |
330 |
0.56 |
| chr8_105225257_105225974 | 0.27 |
D230025D16Rik |
RIKEN cDNA D230025D16 gene |
348 |
0.74 |
| chr9_50658697_50660266 | 0.27 |
Dlat |
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
299 |
0.84 |
| chr8_11009176_11009974 | 0.26 |
Irs2 |
insulin receptor substrate 2 |
1117 |
0.36 |
| chr14_52196647_52197712 | 0.26 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
237 |
0.56 |
| chr4_32799944_32801043 | 0.26 |
Lyrm2 |
LYR motif containing 2 |
211 |
0.94 |
| chr6_147042566_147042769 | 0.26 |
Mrps35 |
mitochondrial ribosomal protein S35 |
97 |
0.95 |
| chr3_95871268_95871981 | 0.26 |
C920021L13Rik |
RIKEN cDNA C920021L13 gene |
93 |
0.5 |
| chr7_4865726_4866166 | 0.25 |
Isoc2b |
isochorismatase domain containing 2b |
222 |
0.84 |
| chr2_180545211_180545584 | 0.25 |
Mrgbp |
MRG/MORF4L binding protein |
35907 |
0.1 |
| chr12_84970237_84970929 | 0.25 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
241 |
0.56 |
| chr17_15395813_15397019 | 0.25 |
Fam120b |
family with sequence similarity 120, member B |
168 |
0.94 |
| chr5_25529908_25530568 | 0.24 |
1700096K18Rik |
RIKEN cDNA 1700096K18 gene |
225 |
0.88 |
| chr8_126591197_126592184 | 0.24 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
2296 |
0.34 |
| chr15_99701831_99703358 | 0.24 |
Smarcd1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
0 |
0.93 |
| chrX_56156268_56156478 | 0.24 |
Gm14639 |
predicted gene 14639 |
20 |
0.97 |
| chr7_45922758_45923210 | 0.24 |
Ccdc114 |
coiled-coil domain containing 114 |
1088 |
0.21 |
| chr13_36117883_36118106 | 0.24 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
114 |
0.84 |
| chr7_44853717_44855163 | 0.24 |
Akt1s1 |
AKT1 substrate 1 (proline-rich) |
1869 |
0.12 |
| chr5_140387535_140387686 | 0.23 |
Snx8 |
sorting nexin 8 |
1631 |
0.28 |
| chr5_115559545_115560657 | 0.23 |
Rplp0 |
ribosomal protein, large, P0 |
478 |
0.66 |
| chr7_29859641_29859994 | 0.23 |
Zfp420 |
zinc finger protein 420 |
162 |
0.89 |
| chr9_72791719_72792022 | 0.23 |
Prtg |
protogenin |
15004 |
0.11 |
| chr9_62979878_62980820 | 0.23 |
Pias1 |
protein inhibitor of activated STAT 1 |
530 |
0.8 |
| chr1_57970048_57970402 | 0.23 |
Kctd18 |
potassium channel tetramerisation domain containing 18 |
97 |
0.97 |
| chr9_107571266_107571916 | 0.23 |
Hyal2 |
hyaluronoglucosaminidase 2 |
381 |
0.57 |
| chr11_117344543_117345735 | 0.23 |
Septin9 |
septin 9 |
8041 |
0.2 |
| chr5_134183780_134184412 | 0.23 |
Gtf2ird2 |
GTF2I repeat domain containing 2 |
69 |
0.96 |
| chr2_167656344_167657144 | 0.23 |
Gm20431 |
predicted gene 20431 |
4752 |
0.11 |
| chr9_96891577_96891728 | 0.23 |
Pxylp1 |
2-phosphoxylose phosphatase 1 |
438 |
0.78 |
| chr10_44852553_44852796 | 0.22 |
Gm6983 |
predicted gene 6983 |
60 |
0.97 |
| chr4_116073957_116074118 | 0.22 |
Uqcrh |
ubiquinol-cytochrome c reductase hinge protein |
994 |
0.31 |
| chr7_80947492_80948211 | 0.22 |
Sec11a |
SEC11 homolog A, signal peptidase complex subunit |
71 |
0.95 |
| chr2_30061841_30063386 | 0.22 |
Set |
SET nuclear oncogene |
12 |
0.96 |
| chr2_119115391_119115542 | 0.22 |
Rad51 |
RAD51 recombinase |
2649 |
0.18 |
| chr15_5116689_5117882 | 0.22 |
Rpl37 |
ribosomal protein L37 |
1 |
0.94 |
| chrX_50590880_50591432 | 0.22 |
Firre |
functional intergenic repeating RNA element |
803 |
0.72 |
| chr7_126474488_126475011 | 0.22 |
Sh2b1 |
SH2B adaptor protein 1 |
73 |
0.94 |
| chr4_124714467_124714687 | 0.22 |
Sf3a3 |
splicing factor 3a, subunit 3 |
199 |
0.86 |
| chr17_35969746_35970067 | 0.21 |
Abcf1 |
ATP-binding cassette, sub-family F (GCN20), member 1 |
145 |
0.86 |
| chrX_57325690_57326987 | 0.21 |
Arhgef6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
261 |
0.93 |
| chr8_83998330_83998685 | 0.21 |
Samd1 |
sterile alpha motif domain containing 1 |
83 |
0.91 |
| chr8_124899499_124899768 | 0.21 |
Gm45709 |
predicted gene 45709 |
497 |
0.62 |
| chr8_46470840_46472147 | 0.21 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
310 |
0.85 |
| chr15_98144892_98145974 | 0.21 |
Asb8 |
ankyrin repeat and SOCS box-containing 8 |
226 |
0.89 |
| chr13_4191088_4191744 | 0.21 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
232 |
0.89 |
| chr2_119112294_119112797 | 0.21 |
Rad51 |
RAD51 recombinase |
248 |
0.88 |
| chr5_120116535_120116888 | 0.21 |
Rbm19 |
RNA binding motif protein 19 |
191 |
0.95 |
| chr19_6856906_6858230 | 0.21 |
Ccdc88b |
coiled-coil domain containing 88B |
643 |
0.54 |
| chr1_156035357_156035647 | 0.21 |
Tor1aip2 |
torsin A interacting protein 2 |
99 |
0.87 |
| chr11_70763963_70764993 | 0.21 |
Zfp3 |
zinc finger protein 3 |
269 |
0.79 |
| chr10_82984904_82986571 | 0.21 |
Chst11 |
carbohydrate sulfotransferase 11 |
239 |
0.93 |
| chr14_32513251_32514390 | 0.20 |
Ercc6 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 |
290 |
0.89 |
| chr3_28171238_28171870 | 0.20 |
Gm42196 |
predicted gene, 42196 |
4999 |
0.26 |
| chr14_51064866_51065193 | 0.20 |
Olfr750 |
olfactory receptor 750 |
6413 |
0.08 |
| chr14_24487009_24487267 | 0.20 |
Rps24 |
ribosomal protein S24 |
13 |
0.55 |
| chr17_36270914_36272068 | 0.20 |
Trim39 |
tripartite motif-containing 39 |
76 |
0.91 |
| chr17_28234245_28235051 | 0.20 |
Ppard |
peroxisome proliferator activator receptor delta |
1841 |
0.22 |
| chr8_125013082_125013762 | 0.20 |
Tsnax |
translin-associated factor X |
373 |
0.83 |
| chr2_167421218_167421702 | 0.20 |
Slc9a8 |
solute carrier family 9 (sodium/hydrogen exchanger), member 8 |
252 |
0.92 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.5 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.4 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.4 | GO:0061620 | glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 0.2 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.6 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 0.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.2 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.3 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:0031506 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.5 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.1 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.0 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) |
| 0.0 | 0.2 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.0 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0048538 | thymus development(GO:0048538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.2 | GO:0016428 | tRNA (cytosine) methyltransferase activity(GO:0016427) tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0018449 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 1.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0045543 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |