Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
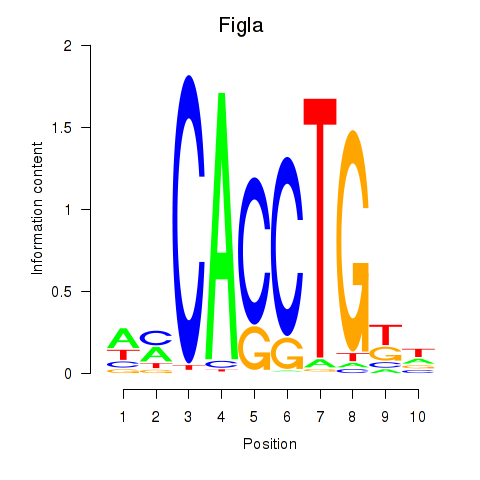
Results for Figla
Z-value: 0.31
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSMUSG00000030001.3 | folliculogenesis specific basic helix-loop-helix |
Activity of the Figla motif across conditions
Conditions sorted by the z-value of the Figla motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
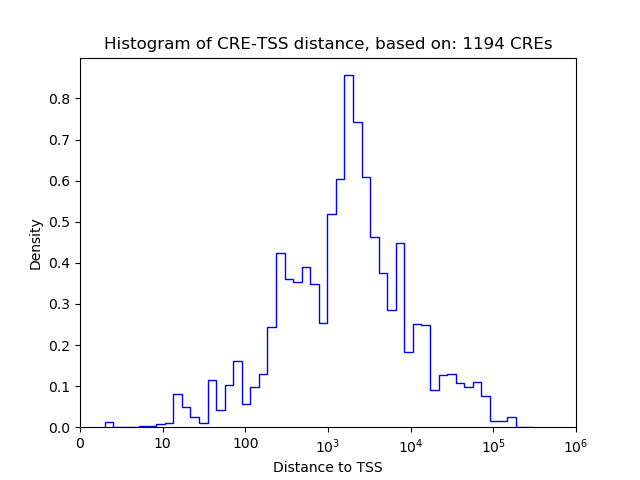
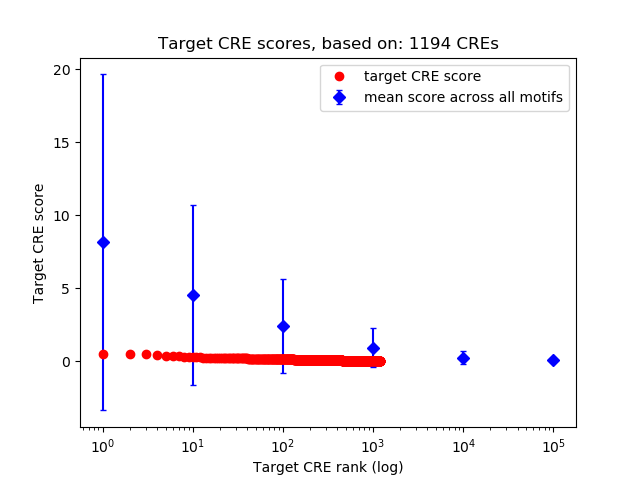
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_16100111_16100955 | 0.48 |
Napa |
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
1895 |
0.23 |
| chr11_96946068_96946479 | 0.48 |
D030028A08Rik |
RIKEN cDNA D030028A08 gene |
1961 |
0.15 |
| chr4_133872481_133873035 | 0.47 |
Gm12977 |
predicted gene 12977 |
276 |
0.47 |
| chr11_102357545_102358717 | 0.44 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
2437 |
0.16 |
| chr5_134915097_134915436 | 0.36 |
Cldn13 |
claudin 13 |
260 |
0.81 |
| chr12_105038279_105038596 | 0.35 |
Glrx5 |
glutaredoxin 5 |
3221 |
0.13 |
| chr11_11732922_11733382 | 0.35 |
Gm12000 |
predicted gene 12000 |
36713 |
0.13 |
| chr17_57234704_57235202 | 0.31 |
C3 |
complement component 3 |
6817 |
0.11 |
| chr18_32555746_32555897 | 0.29 |
Gypc |
glycophorin C |
4159 |
0.24 |
| chr8_94169527_94169694 | 0.29 |
Mt2 |
metallothionein 2 |
3054 |
0.13 |
| chr5_113973724_113975031 | 0.27 |
Ssh1 |
slingshot protein phosphatase 1 |
7621 |
0.13 |
| chr12_105034788_105035573 | 0.27 |
Glrx5 |
glutaredoxin 5 |
36 |
0.95 |
| chr9_120114964_120115764 | 0.26 |
Slc25a38 |
solute carrier family 25, member 38 |
375 |
0.69 |
| chr9_64807169_64807758 | 0.25 |
Dennd4a |
DENN/MADD domain containing 4A |
3877 |
0.25 |
| chr13_73468941_73470244 | 0.25 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
2159 |
0.35 |
| chr4_87881641_87881792 | 0.25 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
7174 |
0.3 |
| chr10_58373981_58374132 | 0.25 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
2602 |
0.27 |
| chr13_9095816_9096692 | 0.24 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
2272 |
0.25 |
| chr3_127893429_127894301 | 0.24 |
Fam241a |
family with sequence similarity 241, member A |
2423 |
0.2 |
| chr15_80743676_80743993 | 0.24 |
Gm49512 |
predicted gene, 49512 |
31563 |
0.13 |
| chr11_102363631_102364272 | 0.24 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
247 |
0.85 |
| chr6_86547683_86548400 | 0.23 |
1600020E01Rik |
RIKEN cDNA 1600020E01 gene |
60 |
0.95 |
| chr7_127090806_127091953 | 0.22 |
AI467606 |
expressed sequence AI467606 |
20 |
0.93 |
| chr14_75178051_75179727 | 0.22 |
Lcp1 |
lymphocyte cytosolic protein 1 |
2681 |
0.23 |
| chr5_143819014_143819653 | 0.22 |
Eif2ak1 |
eukaryotic translation initiation factor 2 alpha kinase 1 |
1386 |
0.38 |
| chr15_83169748_83171160 | 0.22 |
Cyb5r3 |
cytochrome b5 reductase 3 |
52 |
0.95 |
| chr16_91804661_91805209 | 0.22 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
299 |
0.9 |
| chr7_121073211_121073362 | 0.22 |
Igsf6 |
immunoglobulin superfamily, member 6 |
1286 |
0.24 |
| chr2_109281814_109282372 | 0.22 |
Kif18a |
kinesin family member 18A |
1224 |
0.4 |
| chr12_84150828_84152588 | 0.22 |
Pnma1 |
paraneoplastic antigen MA1 |
3219 |
0.13 |
| chr8_105300231_105300842 | 0.21 |
E2f4 |
E2F transcription factor 4 |
2819 |
0.08 |
| chr2_93455051_93455258 | 0.20 |
Gm10804 |
predicted gene 10804 |
2333 |
0.24 |
| chr12_103656514_103657055 | 0.20 |
Serpina6 |
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
318 |
0.81 |
| chr11_4122925_4123415 | 0.20 |
Sec14l2 |
SEC14-like lipid binding 2 |
245 |
0.83 |
| chr3_83008148_83008803 | 0.20 |
Gm30097 |
predicted gene, 30097 |
13 |
0.92 |
| chr6_57754218_57754905 | 0.20 |
Lancl2 |
LanC (bacterial lantibiotic synthetase component C)-like 2 |
51474 |
0.08 |
| chr8_88298512_88298878 | 0.19 |
Adcy7 |
adenylate cyclase 7 |
1684 |
0.37 |
| chrX_101275149_101275321 | 0.19 |
Med12 |
mediator complex subunit 12 |
975 |
0.34 |
| chr1_80030817_80031190 | 0.19 |
Gm28058 |
predicted gene 28058 |
67619 |
0.09 |
| chr2_45055787_45056274 | 0.19 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
1789 |
0.4 |
| chr10_81351824_81352381 | 0.19 |
Hmg20b |
high mobility group 20B |
1622 |
0.13 |
| chr3_146407041_146407816 | 0.19 |
Ssx2ip |
synovial sarcoma, X 2 interacting protein |
2450 |
0.2 |
| chr2_163547257_163548345 | 0.19 |
Hnf4a |
hepatic nuclear factor 4, alpha |
613 |
0.63 |
| chr17_79926076_79926433 | 0.19 |
Gm6552 |
predicted gene 6552 |
8318 |
0.18 |
| chr9_63760744_63760895 | 0.19 |
Smad3 |
SMAD family member 3 |
2825 |
0.3 |
| chr4_46396091_46396440 | 0.18 |
Trmo |
tRNA methyltransferase O |
6828 |
0.13 |
| chr1_45872040_45872606 | 0.18 |
Gm5526 |
predicted pseudogene 5526 |
14569 |
0.12 |
| chr4_6194375_6194526 | 0.18 |
Ubxn2b |
UBX domain protein 2B |
3352 |
0.3 |
| chr7_132778272_132778793 | 0.18 |
Fam53b |
family with sequence similarity 53, member B |
1616 |
0.39 |
| chr18_64504795_64505332 | 0.18 |
Nars |
asparaginyl-tRNA synthetase |
13 |
0.97 |
| chr17_46159788_46160011 | 0.18 |
Gtpbp2 |
GTP binding protein 2 |
1133 |
0.32 |
| chr6_127578287_127578988 | 0.18 |
Cracr2a |
calcium release activated channel regulator 2A |
662 |
0.73 |
| chr12_81028018_81028551 | 0.18 |
Smoc1 |
SPARC related modular calcium binding 1 |
1456 |
0.42 |
| chr10_40149812_40150388 | 0.18 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
7842 |
0.13 |
| chr3_51416101_51417305 | 0.17 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
89 |
0.94 |
| chr9_108083459_108083743 | 0.17 |
Rnf123 |
ring finger protein 123 |
255 |
0.67 |
| chr15_57891181_57891332 | 0.17 |
Derl1 |
Der1-like domain family, member 1 |
862 |
0.63 |
| chr6_118757250_118757706 | 0.17 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
496 |
0.86 |
| chr17_71206427_71206897 | 0.17 |
Lpin2 |
lipin 2 |
1986 |
0.3 |
| chr15_80744676_80744939 | 0.17 |
Gm49512 |
predicted gene, 49512 |
32536 |
0.13 |
| chr7_45522122_45522472 | 0.16 |
Ppp1r15a |
protein phosphatase 1, regulatory subunit 15A |
1472 |
0.15 |
| chr6_29698375_29698642 | 0.16 |
Tspan33 |
tetraspanin 33 |
4274 |
0.2 |
| chr15_86104572_86104788 | 0.16 |
Gm15722 |
predicted gene 15722 |
14620 |
0.17 |
| chr13_91461118_91462460 | 0.16 |
Ssbp2 |
single-stranded DNA binding protein 2 |
608 |
0.81 |
| chr19_39739090_39739575 | 0.16 |
Cyp2c68 |
cytochrome P450, family 2, subfamily c, polypeptide 68 |
1722 |
0.45 |
| chr12_76256073_76256502 | 0.16 |
Mthfd1 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
807 |
0.38 |
| chr4_43612409_43612615 | 0.16 |
Gm25262 |
predicted gene, 25262 |
18913 |
0.06 |
| chr12_75320470_75320850 | 0.16 |
Rhoj |
ras homolog family member J |
11779 |
0.27 |
| chr6_38920967_38921907 | 0.16 |
Tbxas1 |
thromboxane A synthase 1, platelet |
2413 |
0.31 |
| chr13_45546862_45547471 | 0.16 |
Gmpr |
guanosine monophosphate reductase |
1013 |
0.64 |
| chr18_67241729_67242343 | 0.16 |
Mppe1 |
metallophosphoesterase 1 |
3241 |
0.19 |
| chr17_84180639_84182724 | 0.16 |
Gm36279 |
predicted gene, 36279 |
4075 |
0.18 |
| chr8_105941562_105942262 | 0.16 |
Lcat |
lecithin cholesterol acyltransferase |
1470 |
0.16 |
| chr2_170427280_170427591 | 0.16 |
Bcas1 |
breast carcinoma amplified sequence 1 |
393 |
0.83 |
| chr4_139335404_139335655 | 0.16 |
AL807811.1 |
aldo-keto reductase family 7 member A2 (AKR7A2) pseudogene |
1651 |
0.17 |
| chr2_84738253_84739261 | 0.16 |
Mir130a |
microRNA 130a |
2421 |
0.12 |
| chr17_27558079_27558612 | 0.16 |
Hmga1 |
high mobility group AT-hook 1 |
1650 |
0.16 |
| chr16_90987984_90989157 | 0.16 |
Gm15965 |
predicted gene 15965 |
4663 |
0.12 |
| chr13_51647699_51648388 | 0.16 |
Gm22806 |
predicted gene, 22806 |
156 |
0.94 |
| chr10_53598517_53599274 | 0.16 |
Asf1a |
anti-silencing function 1A histone chaperone |
1138 |
0.28 |
| chr6_120819964_120820819 | 0.16 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
1992 |
0.25 |
| chr7_75863523_75863976 | 0.15 |
Klhl25 |
kelch-like 25 |
15308 |
0.22 |
| chr15_80079782_80080738 | 0.15 |
Gm22077 |
predicted gene, 22077 |
83 |
0.67 |
| chr8_122719956_122721464 | 0.15 |
C230057M02Rik |
RIKEN cDNA C230057M02 gene |
17801 |
0.09 |
| chr5_66080287_66081072 | 0.15 |
Rbm47 |
RNA binding motif protein 47 |
305 |
0.84 |
| chr18_84884692_84885054 | 0.15 |
Cyb5a |
cytochrome b5 type A (microsomal) |
7272 |
0.17 |
| chr11_55468160_55468311 | 0.15 |
G3bp1 |
GTPase activating protein (SH3 domain) binding protein 1 |
1450 |
0.26 |
| chr10_81376905_81377216 | 0.15 |
Fzr1 |
fizzy and cell division cycle 20 related 1 |
1320 |
0.16 |
| chr4_118568999_118569601 | 0.15 |
Tmem125 |
transmembrane protein 125 |
25256 |
0.1 |
| chr17_48466649_48466831 | 0.15 |
Unc5cl |
unc-5 family C-terminal like |
4336 |
0.16 |
| chr19_55939739_55939984 | 0.15 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
41552 |
0.17 |
| chr10_80117771_80117922 | 0.15 |
Stk11 |
serine/threonine kinase 11 |
1492 |
0.21 |
| chr13_21751920_21752638 | 0.15 |
Gm44454 |
predicted gene, 44454 |
1166 |
0.14 |
| chr2_4572310_4572962 | 0.15 |
Frmd4a |
FERM domain containing 4A |
6460 |
0.21 |
| chr11_78832870_78833569 | 0.15 |
Lyrm9 |
LYR motif containing 9 |
6590 |
0.18 |
| chr12_76907283_76907638 | 0.15 |
Fntb |
farnesyltransferase, CAAX box, beta |
2026 |
0.3 |
| chr8_107586529_107586949 | 0.14 |
Psmd7 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
1719 |
0.37 |
| chr6_83056416_83056567 | 0.14 |
Aup1 |
ancient ubiquitous protein 1 |
21 |
0.89 |
| chrX_9748838_9749171 | 0.14 |
Dynlt3 |
dynein light chain Tctex-type 3 |
86001 |
0.08 |
| chr17_5492680_5493910 | 0.14 |
Zdhhc14 |
zinc finger, DHHC domain containing 14 |
738 |
0.66 |
| chr12_24892833_24893055 | 0.14 |
Mboat2 |
membrane bound O-acyltransferase domain containing 2 |
61313 |
0.09 |
| chr5_22342338_22343153 | 0.14 |
Reln |
reelin |
1945 |
0.26 |
| chr18_56922535_56922923 | 0.14 |
Marchf3 |
membrane associated ring-CH-type finger 3 |
2786 |
0.27 |
| chr15_80257680_80258950 | 0.14 |
Atf4 |
activating transcription factor 4 |
2792 |
0.15 |
| chr13_3863108_3863464 | 0.14 |
Calm5 |
calmodulin 5 |
9018 |
0.12 |
| chr19_6300669_6301196 | 0.14 |
Ehd1 |
EH-domain containing 1 |
3018 |
0.1 |
| chr8_120490793_120490944 | 0.14 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
2421 |
0.23 |
| chr2_181286147_181286410 | 0.14 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
796 |
0.45 |
| chr9_106156833_106157412 | 0.14 |
Glyctk |
glycerate kinase |
128 |
0.86 |
| chr11_78180253_78181436 | 0.14 |
Snord42a |
small nucleolar RNA, C/D box 42A |
519 |
0.33 |
| chr5_27790098_27791663 | 0.13 |
Paxip1 |
PAX interacting (with transcription-activation domain) protein 1 |
503 |
0.77 |
| chr4_141751491_141751738 | 0.13 |
Agmat |
agmatine ureohydrolase (agmatinase) |
4942 |
0.14 |
| chr10_111476556_111477031 | 0.13 |
Nap1l1 |
nucleosome assembly protein 1-like 1 |
3512 |
0.17 |
| chr11_60942458_60942611 | 0.13 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
654 |
0.63 |
| chr2_118658075_118658340 | 0.13 |
Pak6 |
p21 (RAC1) activated kinase 6 |
5096 |
0.16 |
| chr9_72408510_72409012 | 0.13 |
Gm27255 |
predicted gene 27255 |
507 |
0.52 |
| chr5_146891291_146891493 | 0.13 |
Rasl11a |
RAS-like, family 11, member A |
45627 |
0.09 |
| chr11_6295096_6295368 | 0.13 |
Ogdh |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
2627 |
0.17 |
| chr7_141392818_141393024 | 0.13 |
Taldo1 |
transaldolase 1 |
678 |
0.42 |
| chr11_52076244_52076395 | 0.13 |
Ppp2ca |
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
22362 |
0.11 |
| chr4_134869303_134870057 | 0.13 |
Rhd |
Rh blood group, D antigen |
5144 |
0.18 |
| chr8_122698741_122699314 | 0.13 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
82 |
0.93 |
| chr11_95805344_95805600 | 0.13 |
Phospho1 |
phosphatase, orphan 1 |
19027 |
0.1 |
| chr13_73474220_73474371 | 0.13 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
2544 |
0.32 |
| chr8_69901818_69903173 | 0.13 |
Ndufa13 |
NADH:ubiquinone oxidoreductase subunit A13 |
63 |
0.61 |
| chr4_63561098_63561515 | 0.13 |
Tmem268 |
transmembrane protein 268 |
946 |
0.42 |
| chr2_161066341_161066692 | 0.13 |
Chd6 |
chromodomain helicase DNA binding protein 6 |
13938 |
0.19 |
| chr10_75882493_75882971 | 0.12 |
Gm47744 |
predicted gene, 47744 |
7301 |
0.09 |
| chr5_28457868_28460972 | 0.12 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
1146 |
0.51 |
| chr1_134221026_134221177 | 0.12 |
Adora1 |
adenosine A1 receptor |
13477 |
0.13 |
| chr5_30668984_30669165 | 0.12 |
Cenpa |
centromere protein A |
2076 |
0.22 |
| chr2_105125289_105128976 | 0.12 |
Wt1 |
Wilms tumor 1 homolog |
78 |
0.91 |
| chr13_45507294_45508710 | 0.12 |
Gmpr |
guanosine monophosphate reductase |
558 |
0.8 |
| chr6_67035838_67036542 | 0.12 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
409 |
0.66 |
| chr7_142995964_142996137 | 0.12 |
Tspan32os |
tetraspanin 32, opposite strand |
5272 |
0.14 |
| chr17_26199126_26199427 | 0.12 |
Pdia2 |
protein disulfide isomerase associated 2 |
189 |
0.84 |
| chr1_179853722_179853937 | 0.12 |
Ahctf1 |
AT hook containing transcription factor 1 |
50149 |
0.12 |
| chr9_106888249_106888666 | 0.12 |
Rbm15b |
RNA binding motif protein 15B |
1029 |
0.34 |
| chr17_71183626_71184775 | 0.12 |
Lpin2 |
lipin 2 |
222 |
0.92 |
| chr3_88500857_88501223 | 0.12 |
Lmna |
lamin A |
2267 |
0.13 |
| chr9_72448745_72450029 | 0.12 |
Gm27231 |
predicted gene 27231 |
7608 |
0.08 |
| chr9_71484793_71485892 | 0.12 |
Polr2m |
polymerase (RNA) II (DNA directed) polypeptide M |
384 |
0.9 |
| chr15_73179775_73180371 | 0.12 |
Ago2 |
argonaute RISC catalytic subunit 2 |
2226 |
0.31 |
| chrX_13202535_13203215 | 0.12 |
Rpl3-ps1 |
ribosomal protein L3, pseudogene 1 |
304 |
0.82 |
| chr2_26474742_26475391 | 0.12 |
Notch1 |
notch 1 |
4568 |
0.11 |
| chr3_90429599_90429750 | 0.12 |
Ints3 |
integrator complex subunit 3 |
3629 |
0.12 |
| chr18_84854906_84855693 | 0.11 |
Cyb5a |
cytochrome b5 type A (microsomal) |
330 |
0.86 |
| chr1_6730862_6731309 | 0.11 |
St18 |
suppression of tumorigenicity 18 |
983 |
0.68 |
| chr1_52231983_52233150 | 0.11 |
Gls |
glutaminase |
245 |
0.93 |
| chr1_20818287_20818966 | 0.11 |
Mcm3 |
minichromosome maintenance complex component 3 |
1634 |
0.25 |
| chr2_119619277_119620230 | 0.11 |
Nusap1 |
nucleolar and spindle associated protein 1 |
1024 |
0.3 |
| chr10_80859175_80859326 | 0.11 |
Sppl2b |
signal peptide peptidase like 2B |
2129 |
0.13 |
| chr13_99754101_99754292 | 0.11 |
Gm24471 |
predicted gene, 24471 |
61667 |
0.12 |
| chr17_25225774_25226523 | 0.11 |
Unkl |
unkempt family like zinc finger |
3369 |
0.1 |
| chr3_88953114_88953265 | 0.11 |
Dap3 |
death associated protein 3 |
2008 |
0.18 |
| chr1_180817810_180817961 | 0.11 |
H3f3a |
H3.3 histone A |
3942 |
0.12 |
| chr14_31431914_31432260 | 0.11 |
Sh3bp5 |
SH3-domain binding protein 5 (BTK-associated) |
3978 |
0.17 |
| chr5_64807638_64809344 | 0.11 |
Klf3 |
Kruppel-like factor 3 (basic) |
3848 |
0.17 |
| chr14_31260741_31261456 | 0.11 |
Bap1 |
Brca1 associated protein 1 |
3607 |
0.12 |
| chr9_110699580_110699731 | 0.11 |
Ccdc12 |
coiled-coil domain containing 12 |
10239 |
0.11 |
| chr17_49429375_49429751 | 0.11 |
Gm20540 |
predicted gene 20540 |
323 |
0.83 |
| chr11_69254981_69255132 | 0.11 |
Gm23194 |
predicted gene, 23194 |
14457 |
0.09 |
| chr10_127730237_127730758 | 0.11 |
Tac2 |
tachykinin 2 |
2650 |
0.12 |
| chr17_32117457_32118702 | 0.11 |
Gm17276 |
predicted gene, 17276 |
4332 |
0.15 |
| chr14_8258997_8259161 | 0.11 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
247 |
0.93 |
| chr2_174296925_174297566 | 0.11 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
614 |
0.6 |
| chr4_125727107_125727874 | 0.11 |
2610028E06Rik |
RIKEN cDNA 2610028E06 gene |
171492 |
0.03 |
| chr13_98890050_98891071 | 0.11 |
Tnpo1 |
transportin 1 |
438 |
0.78 |
| chr4_141161340_141161491 | 0.11 |
Fbxo42 |
F-box protein 42 |
13493 |
0.11 |
| chr13_95698558_95698709 | 0.11 |
F2rl2 |
coagulation factor II (thrombin) receptor-like 2 |
1780 |
0.27 |
| chr16_24199309_24199754 | 0.11 |
Gm31814 |
predicted gene, 31814 |
16715 |
0.19 |
| chr18_88758573_88758778 | 0.11 |
Socs6 |
suppressor of cytokine signaling 6 |
184 |
0.96 |
| chr11_44528978_44529659 | 0.11 |
Rnf145 |
ring finger protein 145 |
9013 |
0.18 |
| chr7_128689234_128689975 | 0.11 |
Gm43580 |
predicted gene 43580 |
1069 |
0.26 |
| chr19_7240047_7241153 | 0.11 |
Naa40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
434 |
0.7 |
| chr4_108961053_108961426 | 0.11 |
8030443G20Rik |
RIKEN cDNA 8030443G20 gene |
10838 |
0.13 |
| chr13_49306649_49306800 | 0.11 |
Fgd3 |
FYVE, RhoGEF and PH domain containing 3 |
2516 |
0.28 |
| chr4_126260482_126261460 | 0.11 |
Trappc3 |
trafficking protein particle complex 3 |
1354 |
0.3 |
| chr15_89607900_89608068 | 0.11 |
Rabl2 |
RAB, member RAS oncogene family-like 2 |
16061 |
0.15 |
| chr4_46413029_46413416 | 0.11 |
Hemgn |
hemogen |
284 |
0.86 |
| chr7_3666562_3667626 | 0.11 |
Tmc4 |
transmembrane channel-like gene family 4 |
211 |
0.74 |
| chr6_124832302_124832570 | 0.11 |
Cdca3 |
cell division cycle associated 3 |
2061 |
0.12 |
| chr12_30905934_30906220 | 0.11 |
Acp1 |
acid phosphatase 1, soluble |
5472 |
0.19 |
| chr7_24368915_24369193 | 0.11 |
Kcnn4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
1209 |
0.26 |
| chr10_60078294_60078579 | 0.10 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
27783 |
0.16 |
| chr18_34783686_34784162 | 0.10 |
Kdm3b |
KDM3B lysine (K)-specific demethylase 3B |
6819 |
0.13 |
| chr11_101449130_101449504 | 0.10 |
Ifi35 |
interferon-induced protein 35 |
747 |
0.35 |
| chr6_131384992_131385187 | 0.10 |
Ybx3 |
Y box protein 3 |
3349 |
0.18 |
| chr7_99822281_99822623 | 0.10 |
Neu3 |
neuraminidase 3 |
5965 |
0.13 |
| chr11_76088787_76088938 | 0.10 |
Vps53 |
VPS53 GARP complex subunit |
3817 |
0.23 |
| chr11_69588843_69589373 | 0.10 |
Trp53 |
transformation related protein 53 |
413 |
0.61 |
| chr3_96223834_96224156 | 0.10 |
H2bc21 |
H2B clustered histone 21 |
2876 |
0.05 |
| chr4_134019273_134020476 | 0.10 |
Lin28a |
lin-28 homolog A (C. elegans) |
1033 |
0.34 |
| chr17_27559013_27559762 | 0.10 |
Hmga1 |
high mobility group AT-hook 1 |
2692 |
0.11 |
| chr3_35923525_35923989 | 0.10 |
Dcun1d1 |
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
2569 |
0.18 |
| chr8_122378366_122378701 | 0.10 |
Zc3h18 |
zinc finger CCCH-type containing 18 |
1852 |
0.2 |
| chr9_63604142_63604361 | 0.10 |
Aagab |
alpha- and gamma-adaptin binding protein |
1591 |
0.32 |
| chr16_58678686_58678993 | 0.10 |
Cpox |
coproporphyrinogen oxidase |
983 |
0.44 |
| chr15_66809584_66809797 | 0.10 |
Sla |
src-like adaptor |
2903 |
0.26 |
| chr11_97780583_97780734 | 0.10 |
Gm27326 |
predicted gene, 27326 |
382 |
0.6 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |