Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
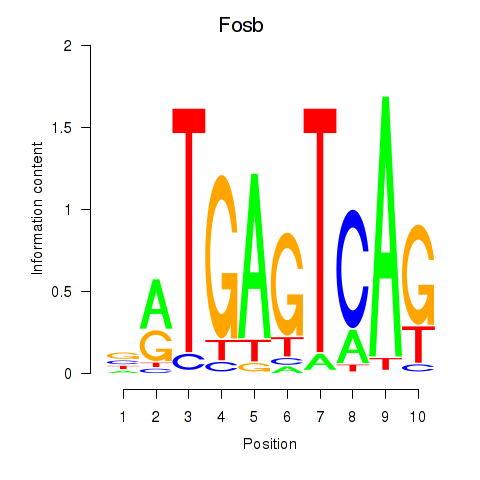
Results for Fosb
Z-value: 0.29
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSMUSG00000003545.2 | FBJ osteosarcoma oncogene B |
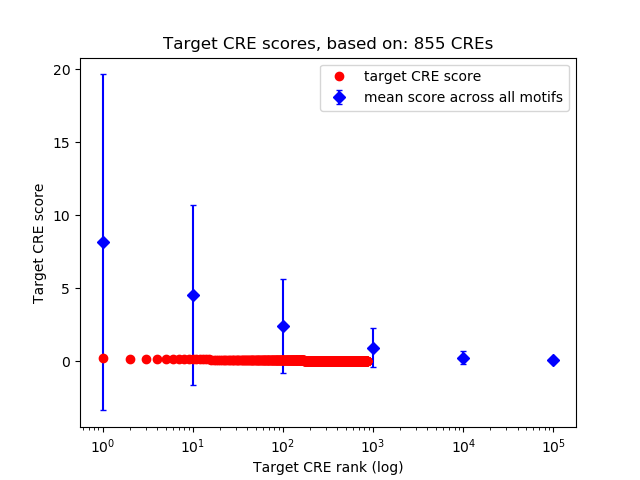
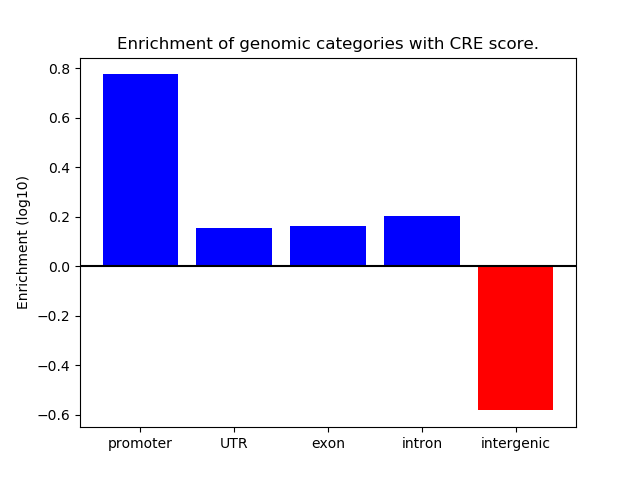
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_19307095_19307459 | Fosb | 2323 | 0.115692 | -0.23 | 7.6e-02 | Click! |
| chr7_19311915_19312066 | Fosb | 1939 | 0.135406 | -0.19 | 1.5e-01 | Click! |
| chr7_19305979_19306638 | Fosb | 3292 | 0.091530 | -0.17 | 2.0e-01 | Click! |
| chr7_19307973_19308124 | Fosb | 1552 | 0.169390 | -0.11 | 3.8e-01 | Click! |
| chr7_19308198_19308579 | Fosb | 1212 | 0.223208 | 0.10 | 4.3e-01 | Click! |
Activity of the Fosb motif across conditions
Conditions sorted by the z-value of the Fosb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
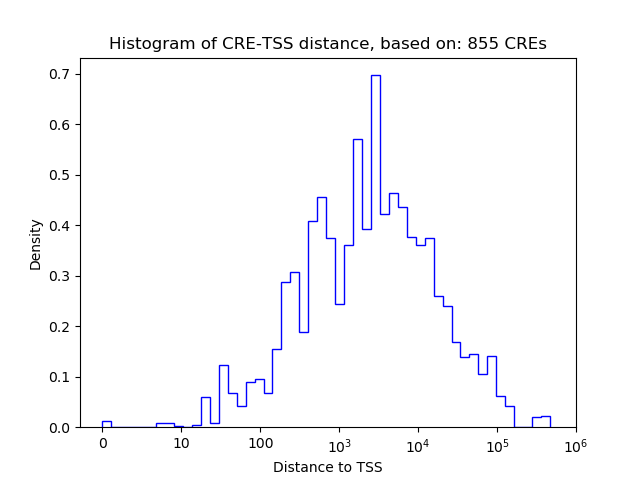
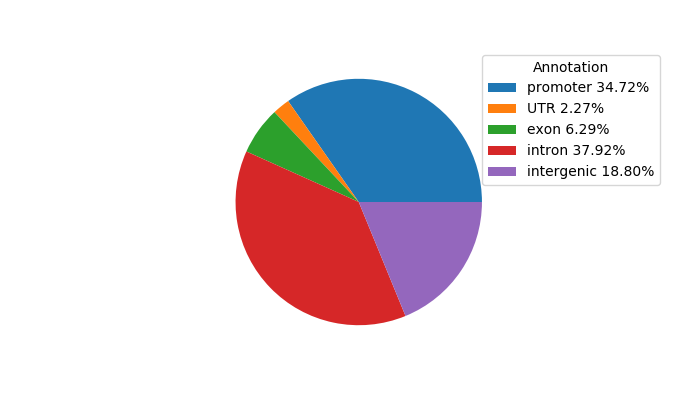
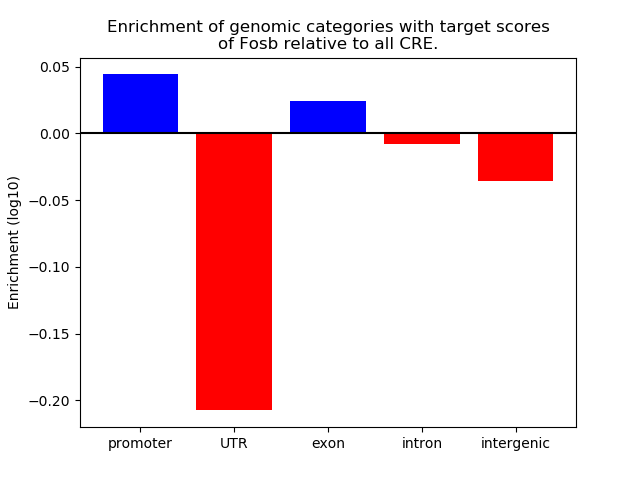
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_117421727_117421878 | 0.25 |
Gm13982 |
predicted gene 13982 |
4105 |
0.31 |
| chr8_125897573_125897839 | 0.19 |
A730098A19Rik |
RIKEN cDNA A730098A19 gene |
208 |
0.8 |
| chr1_165105603_165105901 | 0.18 |
4930568G15Rik |
RIKEN cDNA 4930568G15 gene |
13824 |
0.16 |
| chr6_86082495_86082646 | 0.18 |
Add2 |
adducin 2 (beta) |
211 |
0.91 |
| chr1_75263068_75263858 | 0.16 |
Ptprn |
protein tyrosine phosphatase, receptor type, N |
444 |
0.62 |
| chr15_74194548_74194699 | 0.16 |
Gm15387 |
predicted gene 15387 |
100290 |
0.07 |
| chr10_80070043_80070194 | 0.15 |
Sbno2 |
strawberry notch 2 |
5282 |
0.1 |
| chr9_58847603_58847858 | 0.15 |
Hcn4 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 4 |
24318 |
0.2 |
| chr9_44340100_44340251 | 0.15 |
Hmbs |
hydroxymethylbilane synthase |
459 |
0.53 |
| chr13_73345278_73345429 | 0.15 |
Eprn |
ephemeron, early developmental lncRNA |
2031 |
0.26 |
| chr1_91541582_91541767 | 0.13 |
Asb1 |
ankyrin repeat and SOCS box-containing 1 |
594 |
0.69 |
| chr3_138503819_138503983 | 0.13 |
Gm43725 |
predicted gene 43725 |
311 |
0.82 |
| chrX_7638310_7639997 | 0.13 |
Syp |
synaptophysin |
152 |
0.88 |
| chr18_15345788_15346105 | 0.13 |
A830021F12Rik |
RIKEN cDNA A830021F12 gene |
5988 |
0.22 |
| chr9_65019569_65020184 | 0.12 |
Hacd3 |
3-hydroxyacyl-CoA dehydratase 3 |
1817 |
0.28 |
| chr2_70989412_70989871 | 0.12 |
Mettl8 |
methyltransferase like 8 |
7457 |
0.25 |
| chr18_14654793_14655162 | 0.12 |
Ss18 |
SS18, nBAF chromatin remodeling complex subunit |
3854 |
0.23 |
| chr9_114412717_114412868 | 0.12 |
Glb1 |
galactosidase, beta 1 |
11684 |
0.14 |
| chr6_55338294_55340060 | 0.11 |
Aqp1 |
aquaporin 1 |
2745 |
0.22 |
| chr10_42260699_42260850 | 0.11 |
Foxo3 |
forkhead box O3 |
2408 |
0.38 |
| chr17_25221667_25222954 | 0.11 |
Unkl |
unkempt family like zinc finger |
229 |
0.84 |
| chr1_187910901_187911320 | 0.11 |
Esrrg |
estrogen-related receptor gamma |
86717 |
0.09 |
| chr2_148856545_148856768 | 0.11 |
Cstdc2 |
cystatin domain containing 2 |
5722 |
0.13 |
| chr16_93585766_93585917 | 0.11 |
Setd4 |
SET domain containing 4 |
583 |
0.66 |
| chr5_129955911_129956136 | 0.11 |
Gm43482 |
predicted gene 43482 |
8343 |
0.1 |
| chr12_29529828_29531185 | 0.11 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr6_136659891_136660121 | 0.11 |
Plbd1 |
phospholipase B domain containing 1 |
1867 |
0.3 |
| chr11_87974853_87975012 | 0.11 |
Dynll2 |
dynein light chain LC8-type 2 |
9080 |
0.13 |
| chr18_25745414_25746450 | 0.10 |
Celf4 |
CUGBP, Elav-like family member 4 |
6760 |
0.24 |
| chr2_181767278_181768191 | 0.10 |
Myt1 |
myelin transcription factor 1 |
222 |
0.91 |
| chr3_90694946_90695435 | 0.10 |
S100a9 |
S100 calcium binding protein A9 (calgranulin B) |
521 |
0.62 |
| chr14_31780489_31782000 | 0.10 |
Ankrd28 |
ankyrin repeat domain 28 |
305 |
0.89 |
| chr9_35118981_35119257 | 0.10 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
64 |
0.94 |
| chr13_16009834_16010073 | 0.10 |
Inhba |
inhibin beta-A |
1898 |
0.25 |
| chr10_44539465_44539637 | 0.10 |
Prdm1 |
PR domain containing 1, with ZNF domain |
11050 |
0.15 |
| chr5_69541689_69541957 | 0.10 |
Yipf7 |
Yip1 domain family, member 7 |
766 |
0.59 |
| chrX_152643367_152644550 | 0.10 |
Shroom2 |
shroom family member 2 |
34 |
0.98 |
| chr2_78210324_78210475 | 0.10 |
Gm14461 |
predicted gene 14461 |
27148 |
0.25 |
| chr2_28617937_28618205 | 0.10 |
Gfi1b |
growth factor independent 1B |
2666 |
0.16 |
| chr8_94986231_94987228 | 0.10 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
1161 |
0.36 |
| chr16_41552420_41552571 | 0.10 |
Lsamp |
limbic system-associated membrane protein |
19076 |
0.29 |
| chr1_27884926_27885077 | 0.10 |
Gm5524 |
predicted gene 5524 |
955 |
0.65 |
| chr3_98284074_98284225 | 0.10 |
Gm43189 |
predicted gene 43189 |
2839 |
0.2 |
| chr2_158145151_158146425 | 0.10 |
Tgm2 |
transglutaminase 2, C polypeptide |
578 |
0.71 |
| chr11_79519398_79519813 | 0.10 |
Evi2b |
ecotropic viral integration site 2b |
4157 |
0.12 |
| chr8_123409926_123410387 | 0.09 |
Tubb3 |
tubulin, beta 3 class III |
1268 |
0.19 |
| chrX_75576775_75576926 | 0.09 |
Rab39b |
RAB39B, member RAS oncogene family |
1381 |
0.29 |
| chr7_24485614_24487418 | 0.09 |
Cadm4 |
cell adhesion molecule 4 |
4493 |
0.1 |
| chr2_119299920_119301063 | 0.09 |
Vps18 |
VPS18 CORVET/HOPS core subunit |
11704 |
0.1 |
| chr18_25246238_25246492 | 0.09 |
AW554918 |
expressed sequence AW554918 |
12450 |
0.26 |
| chr4_45949484_45949759 | 0.09 |
Tdrd7 |
tudor domain containing 7 |
15713 |
0.18 |
| chr1_36923512_36923663 | 0.09 |
F830112A20Rik |
RIKEN cDNA F830112A20 gene |
2761 |
0.19 |
| chr3_60531956_60532107 | 0.09 |
Mbnl1 |
muscleblind like splicing factor 1 |
2400 |
0.3 |
| chr8_47027128_47027359 | 0.09 |
4930579M01Rik |
RIKEN cDNA 4930579M01 gene |
10934 |
0.18 |
| chr6_47636667_47636869 | 0.09 |
Gm44141 |
predicted gene, 44141 |
1471 |
0.42 |
| chr3_40745266_40746433 | 0.09 |
Hspa4l |
heat shock protein 4 like |
19 |
0.97 |
| chr10_94919501_94920004 | 0.09 |
Plxnc1 |
plexin C1 |
2905 |
0.28 |
| chr12_41481392_41481887 | 0.09 |
Lrrn3 |
leucine rich repeat protein 3, neuronal |
4792 |
0.26 |
| chr15_76197527_76199931 | 0.09 |
Plec |
plectin |
520 |
0.59 |
| chr1_87620304_87621692 | 0.09 |
Inpp5d |
inositol polyphosphate-5-phosphatase D |
371 |
0.85 |
| chr10_77530297_77531905 | 0.09 |
Itgb2 |
integrin beta 2 |
721 |
0.57 |
| chr3_84563506_84563657 | 0.08 |
Gm38241 |
predicted gene, 38241 |
5794 |
0.17 |
| chr5_118480209_118480360 | 0.08 |
Gm15754 |
predicted gene 15754 |
6683 |
0.21 |
| chr5_88583963_88584847 | 0.08 |
Rufy3 |
RUN and FYVE domain containing 3 |
611 |
0.7 |
| chr5_24839524_24839675 | 0.08 |
Rheb |
Ras homolog enriched in brain |
2829 |
0.22 |
| chr17_87609597_87610246 | 0.08 |
Epcam |
epithelial cell adhesion molecule |
26058 |
0.15 |
| chr9_71722702_71722853 | 0.08 |
Cgnl1 |
cingulin-like 1 |
48783 |
0.13 |
| chr8_69916445_69916596 | 0.08 |
Yjefn3 |
YjeF N-terminal domain containing 3 |
13808 |
0.1 |
| chr18_85908611_85908899 | 0.08 |
Gm5824 |
predicted gene 5824 |
60026 |
0.16 |
| chr10_122387434_122387585 | 0.08 |
Gm36041 |
predicted gene, 36041 |
617 |
0.78 |
| chr2_127140940_127141091 | 0.08 |
Itpripl1 |
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
1575 |
0.32 |
| chr11_80602299_80602557 | 0.08 |
C030013C21Rik |
RIKEN cDNA C030013C21 gene |
93322 |
0.07 |
| chr19_37059612_37059903 | 0.08 |
Gm22714 |
predicted gene, 22714 |
89405 |
0.06 |
| chr11_82845042_82846306 | 0.08 |
Rffl |
ring finger and FYVE like domain containing protein |
418 |
0.75 |
| chr5_148391534_148391736 | 0.08 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
1180 |
0.56 |
| chr4_117412018_117412169 | 0.08 |
Gm12828 |
predicted gene 12828 |
33857 |
0.12 |
| chr14_79535033_79535184 | 0.08 |
Elf1 |
E74-like factor 1 |
19410 |
0.14 |
| chrX_57286626_57286777 | 0.08 |
Arhgef6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
5107 |
0.25 |
| chr7_97456067_97456218 | 0.08 |
Kctd14 |
potassium channel tetramerisation domain containing 14 |
2920 |
0.19 |
| chr11_77800451_77802048 | 0.08 |
Myo18a |
myosin XVIIIA |
49 |
0.97 |
| chr14_122184045_122184304 | 0.08 |
Clybl |
citrate lyase beta like |
2470 |
0.27 |
| chr17_84152977_84153128 | 0.08 |
Gm19696 |
predicted gene, 19696 |
3374 |
0.21 |
| chr6_86666948_86667521 | 0.08 |
Gm44292 |
predicted gene, 44292 |
1683 |
0.2 |
| chr11_60934916_60936338 | 0.08 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
3561 |
0.16 |
| chr9_21917305_21918263 | 0.08 |
Rab3d |
RAB3D, member RAS oncogene family |
305 |
0.79 |
| chr11_88215353_88216424 | 0.08 |
Mrps23 |
mitochondrial ribosomal protein S23 |
11444 |
0.16 |
| chr6_56914111_56914396 | 0.08 |
Gm3793 |
predicted gene 3793 |
7671 |
0.13 |
| chr2_94194378_94194529 | 0.08 |
Gm23630 |
predicted gene, 23630 |
10323 |
0.15 |
| chr7_75571927_75572378 | 0.07 |
Gm44835 |
predicted gene 44835 |
11538 |
0.2 |
| chr4_77743658_77743809 | 0.07 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
468006 |
0.01 |
| chr1_180191356_180191507 | 0.07 |
Coq8a |
coenzyme Q8A |
2052 |
0.25 |
| chr17_72839028_72839198 | 0.07 |
Ypel5 |
yippee like 5 |
742 |
0.76 |
| chr6_50037599_50038047 | 0.07 |
Gm3455 |
predicted gene 3455 |
13363 |
0.26 |
| chr2_136054783_136055114 | 0.07 |
Lamp5 |
lysosomal-associated membrane protein family, member 5 |
2709 |
0.3 |
| chr5_73252277_73252428 | 0.07 |
Fryl |
FRY like transcription coactivator |
4073 |
0.13 |
| chr7_139833633_139836105 | 0.07 |
Adgra1 |
adhesion G protein-coupled receptor A1 |
93 |
0.96 |
| chr5_107874374_107875235 | 0.07 |
Evi5 |
ecotropic viral integration site 5 |
240 |
0.86 |
| chr8_116115911_116116602 | 0.07 |
4930422C21Rik |
RIKEN cDNA 4930422C21 gene |
71113 |
0.13 |
| chr1_171019411_171019562 | 0.07 |
Fcgr4 |
Fc receptor, IgG, low affinity IV |
566 |
0.59 |
| chr5_77092587_77093501 | 0.07 |
Hopxos |
HOP homeobox, opposite strand |
1488 |
0.28 |
| chr3_126661046_126661197 | 0.07 |
Gm43005 |
predicted gene 43005 |
1258 |
0.33 |
| chr12_111420504_111420734 | 0.07 |
Exoc3l4 |
exocyst complex component 3-like 4 |
30 |
0.96 |
| chr6_128888429_128889097 | 0.07 |
Clec2i |
C-type lectin domain family 2, member i |
1165 |
0.25 |
| chr11_32534988_32535158 | 0.07 |
Stk10 |
serine/threonine kinase 10 |
1768 |
0.34 |
| chr14_120292933_120293084 | 0.07 |
Mbnl2 |
muscleblind like splicing factor 2 |
4666 |
0.3 |
| chr6_9440567_9440718 | 0.07 |
Gm35736 |
predicted gene, 35736 |
293876 |
0.01 |
| chr4_46457288_46457543 | 0.07 |
Anp32b |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
6513 |
0.14 |
| chr3_10088030_10088342 | 0.07 |
Gm9833 |
predicted gene 9833 |
76 |
0.96 |
| chr15_62743706_62743857 | 0.07 |
Gm22521 |
predicted gene, 22521 |
49420 |
0.17 |
| chr6_86031081_86032013 | 0.07 |
Add2 |
adducin 2 (beta) |
2800 |
0.16 |
| chr6_136938496_136939168 | 0.07 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
759 |
0.55 |
| chr8_32007143_32007294 | 0.07 |
Nrg1 |
neuregulin 1 |
1888 |
0.48 |
| chr14_16243838_16244030 | 0.07 |
Gm47794 |
predicted gene, 47794 |
583 |
0.41 |
| chr9_85090484_85090658 | 0.07 |
Gm28070 |
predicted gene 28070 |
14194 |
0.25 |
| chr7_110859157_110859397 | 0.07 |
Lyve1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
3676 |
0.19 |
| chr17_8285131_8285282 | 0.07 |
Mpc1 |
mitochondrial pyruvate carrier 1 |
752 |
0.54 |
| chr15_101373527_101373723 | 0.07 |
Gm35853 |
predicted gene, 35853 |
273 |
0.8 |
| chrX_140592629_140592780 | 0.07 |
Tsc22d3 |
TSC22 domain family, member 3 |
7955 |
0.17 |
| chr6_127329826_127330898 | 0.07 |
Gm42458 |
predicted gene 42458 |
4505 |
0.15 |
| chr3_58469818_58470437 | 0.07 |
Tsc22d2 |
TSC22 domain family, member 2 |
52643 |
0.09 |
| chr5_123025578_123025729 | 0.07 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
10315 |
0.08 |
| chr13_16683110_16683261 | 0.07 |
Gm7537 |
predicted gene 7537 |
43695 |
0.19 |
| chr6_87809573_87810010 | 0.07 |
Rab43 |
RAB43, member RAS oncogene family |
34 |
0.93 |
| chr16_55975114_55975265 | 0.07 |
Zbtb11os1 |
zinc finger and BTB domain containing 11, opposite strand 1 |
572 |
0.54 |
| chr19_12475678_12475983 | 0.06 |
Mpeg1 |
macrophage expressed gene 1 |
15051 |
0.1 |
| chr5_140388135_140388290 | 0.06 |
Snx8 |
sorting nexin 8 |
1029 |
0.43 |
| chr5_113985780_113986314 | 0.06 |
Ssh1 |
slingshot protein phosphatase 1 |
3696 |
0.16 |
| chr17_83874481_83874676 | 0.06 |
C430042M11Rik |
RIKEN cDNA C430042M11 gene |
20160 |
0.14 |
| chr7_46053210_46053361 | 0.06 |
Nomo1 |
nodal modulator 1 |
875 |
0.46 |
| chr14_99631270_99631421 | 0.06 |
Gm9298 |
predicted gene 9298 |
679 |
0.73 |
| chr13_5845041_5845796 | 0.06 |
1700016G22Rik |
RIKEN cDNA 1700016G22 gene |
12143 |
0.16 |
| chr2_128427966_128429191 | 0.06 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
504 |
0.77 |
| chr1_95161422_95161573 | 0.06 |
Gm5264 |
predicted gene 5264 |
95313 |
0.08 |
| chr12_84192576_84193882 | 0.06 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
786 |
0.46 |
| chr8_105822112_105822566 | 0.06 |
Ranbp10 |
RAN binding protein 10 |
4866 |
0.1 |
| chr1_177470226_177470377 | 0.06 |
Gm37306 |
predicted gene, 37306 |
2923 |
0.22 |
| chr17_88460728_88460879 | 0.06 |
Foxn2 |
forkhead box N2 |
20028 |
0.17 |
| chr15_80098310_80099146 | 0.06 |
Syngr1 |
synaptogyrin 1 |
858 |
0.41 |
| chr5_72166515_72166666 | 0.06 |
Commd8 |
COMM domain containing 8 |
870 |
0.59 |
| chr11_77808336_77808487 | 0.06 |
Myo18a |
myosin XVIIIA |
3104 |
0.2 |
| chr1_171607147_171607324 | 0.06 |
Ly9 |
lymphocyte antigen 9 |
94 |
0.94 |
| chr17_29093963_29095348 | 0.06 |
1700023B13Rik |
RIKEN cDNA 1700023B13 gene |
194 |
0.82 |
| chr3_151424642_151424793 | 0.06 |
Adgrl4 |
adhesion G protein-coupled receptor L4 |
13170 |
0.26 |
| chr17_33712070_33712674 | 0.06 |
Marchf2 |
membrane associated ring-CH-type finger 2 |
1009 |
0.36 |
| chr1_168163488_168164140 | 0.06 |
Pbx1 |
pre B cell leukemia homeobox 1 |
848 |
0.75 |
| chr17_74670356_74670507 | 0.06 |
Birc6 |
baculoviral IAP repeat-containing 6 |
224 |
0.94 |
| chr18_75362868_75363274 | 0.06 |
Gm20544 |
predicted gene 20544 |
4017 |
0.23 |
| chr5_21702963_21703132 | 0.06 |
Napepld |
N-acyl phosphatidylethanolamine phospholipase D |
1651 |
0.28 |
| chr6_41701642_41702158 | 0.06 |
Kel |
Kell blood group |
2439 |
0.19 |
| chr18_62176067_62177775 | 0.06 |
Adrb2 |
adrenergic receptor, beta 2 |
3038 |
0.24 |
| chr5_57899193_57899365 | 0.06 |
Gm17977 |
predicted gene, 17977 |
455 |
0.73 |
| chrX_41435768_41435921 | 0.06 |
Gria3 |
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
33104 |
0.21 |
| chrX_110813090_110813270 | 0.06 |
Gm44593 |
predicted gene 44593 |
856 |
0.5 |
| chr2_153531388_153531539 | 0.06 |
Nol4l |
nucleolar protein 4-like |
1492 |
0.42 |
| chr13_101540446_101540959 | 0.06 |
Gm47533 |
predicted gene, 47533 |
4504 |
0.19 |
| chr11_4948719_4949803 | 0.06 |
Nefh |
neurofilament, heavy polypeptide |
1197 |
0.38 |
| chr4_129139860_129140186 | 0.06 |
Fndc5 |
fibronectin type III domain containing 5 |
3024 |
0.15 |
| chr6_3397758_3397977 | 0.06 |
Samd9l |
sterile alpha motif domain containing 9-like |
1705 |
0.33 |
| chr14_65374778_65375936 | 0.06 |
Zfp395 |
zinc finger protein 395 |
36 |
0.97 |
| chr4_3929394_3930248 | 0.06 |
Plag1 |
pleiomorphic adenoma gene 1 |
8561 |
0.14 |
| chr7_56052340_56052491 | 0.06 |
Herc2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
1538 |
0.26 |
| chr1_40268223_40269018 | 0.06 |
Il1r1 |
interleukin 1 receptor, type I |
1963 |
0.34 |
| chr17_70985809_70985960 | 0.06 |
Myl12b |
myosin, light chain 12B, regulatory |
4646 |
0.12 |
| chr8_25103571_25103722 | 0.06 |
Gm45745 |
predicted gene 45745 |
1022 |
0.29 |
| chr2_165982847_165983111 | 0.05 |
Platr29 |
pluripotency associated transcript 29 |
2945 |
0.18 |
| chr6_85915077_85916474 | 0.05 |
Tprkb |
Tp53rk binding protein |
18 |
0.55 |
| chr2_173047915_173048486 | 0.05 |
Gm14453 |
predicted gene 14453 |
13620 |
0.13 |
| chr11_65806557_65807031 | 0.05 |
Zkscan6 |
zinc finger with KRAB and SCAN domains 6 |
381 |
0.88 |
| chr11_5846904_5847055 | 0.05 |
Polm |
polymerase (DNA directed), mu |
8963 |
0.11 |
| chr19_36450921_36451072 | 0.05 |
F530104D19Rik |
RIKEN cDNA F530104D19 gene |
161 |
0.95 |
| chr16_46934741_46934892 | 0.05 |
Gm6912 |
predicted gene 6912 |
139382 |
0.05 |
| chr13_108775293_108775893 | 0.05 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
84573 |
0.1 |
| chr16_38432552_38432929 | 0.05 |
Pla1a |
phospholipase A1 member A |
405 |
0.77 |
| chr9_119977341_119978321 | 0.05 |
Csrnp1 |
cysteine-serine-rich nuclear protein 1 |
496 |
0.65 |
| chr12_52508996_52509147 | 0.05 |
Arhgap5 |
Rho GTPase activating protein 5 |
4729 |
0.21 |
| chr19_56388806_56389568 | 0.05 |
Nrap |
nebulin-related anchoring protein |
690 |
0.66 |
| chr12_84449804_84449955 | 0.05 |
Aldh6a1 |
aldehyde dehydrogenase family 6, subfamily A1 |
1069 |
0.35 |
| chr11_101464234_101464439 | 0.05 |
Rnd2 |
Rho family GTPase 2 |
663 |
0.4 |
| chr12_110953614_110953765 | 0.05 |
Ankrd9 |
ankyrin repeat domain 9 |
24566 |
0.1 |
| chr11_72301272_72302549 | 0.05 |
Xaf1 |
XIAP associated factor 1 |
245 |
0.87 |
| chr7_126880230_126880459 | 0.05 |
Taok2 |
TAO kinase 2 |
2615 |
0.09 |
| chr12_65039383_65039555 | 0.05 |
Prpf39 |
pre-mRNA processing factor 39 |
3103 |
0.15 |
| chr1_120549940_120550406 | 0.05 |
Marco |
macrophage receptor with collagenous structure |
45149 |
0.14 |
| chr18_31441284_31441994 | 0.05 |
Gm26658 |
predicted gene, 26658 |
1321 |
0.39 |
| chr14_68522913_68523064 | 0.05 |
Adam7 |
a disintegrin and metallopeptidase domain 7 |
10701 |
0.23 |
| chr4_43968827_43968978 | 0.05 |
Glipr2 |
GLI pathogenesis-related 2 |
11210 |
0.13 |
| chr11_35544219_35545325 | 0.05 |
Slit3 |
slit guidance ligand 3 |
87 |
0.98 |
| chr16_56717610_56718108 | 0.05 |
Tfg |
Trk-fused gene |
409 |
0.87 |
| chr4_43038844_43040271 | 0.05 |
Fam214b |
family with sequence similarity 214, member B |
226 |
0.87 |
| chr9_19319225_19319376 | 0.05 |
Olfr844 |
olfactory receptor 844 |
764 |
0.55 |
| chr7_90042458_90042609 | 0.05 |
Gm44861 |
predicted gene 44861 |
164 |
0.93 |
| chr16_32859333_32859688 | 0.05 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
8828 |
0.14 |
| chr5_124184090_124186568 | 0.05 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
211 |
0.9 |
| chr16_93363663_93363907 | 0.05 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
1544 |
0.35 |
| chr16_94544511_94544675 | 0.05 |
Vps26c |
VPS26 endosomal protein sorting factor C |
17763 |
0.16 |
| chr2_14640980_14641207 | 0.05 |
Gm13268 |
predicted gene 13268 |
13772 |
0.1 |
| chr10_11229681_11229832 | 0.05 |
Gm16577 |
predicted gene 16577 |
16777 |
0.16 |
| chr1_151250581_151250802 | 0.05 |
Gm24402 |
predicted gene, 24402 |
10223 |
0.14 |
| chr2_125116641_125116953 | 0.05 |
Myef2 |
myelin basic protein expression factor 2, repressor |
76 |
0.96 |
| chr13_54610631_54611317 | 0.05 |
Cltb |
clathrin, light polypeptide (Lcb) |
318 |
0.81 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |