Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
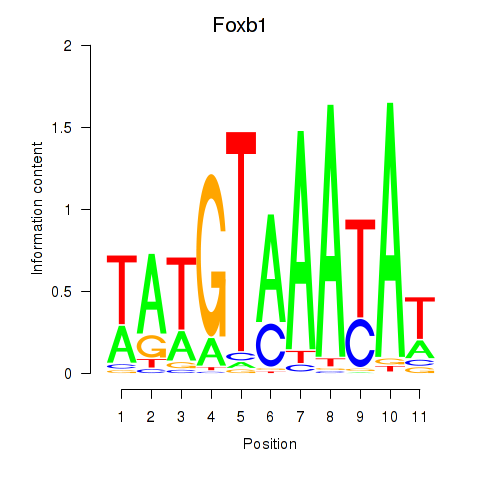
Results for Foxb1
Z-value: 0.79
Transcription factors associated with Foxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxb1
|
ENSMUSG00000059246.4 | forkhead box B1 |
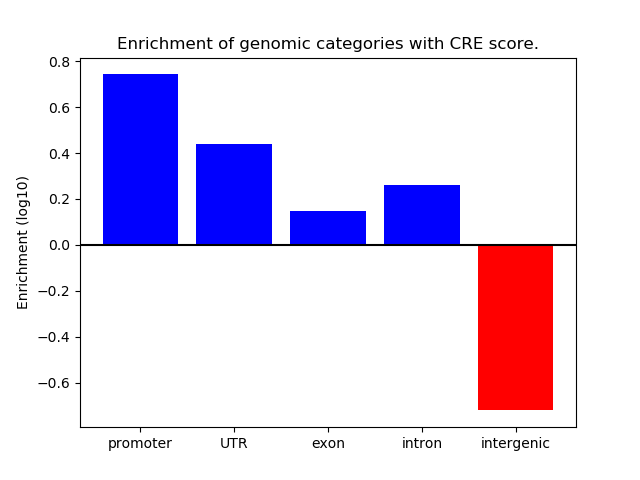
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_69758963_69761490 | Foxb1 | 714 | 0.498655 | 0.51 | 3.8e-05 | Click! |
Activity of the Foxb1 motif across conditions
Conditions sorted by the z-value of the Foxb1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
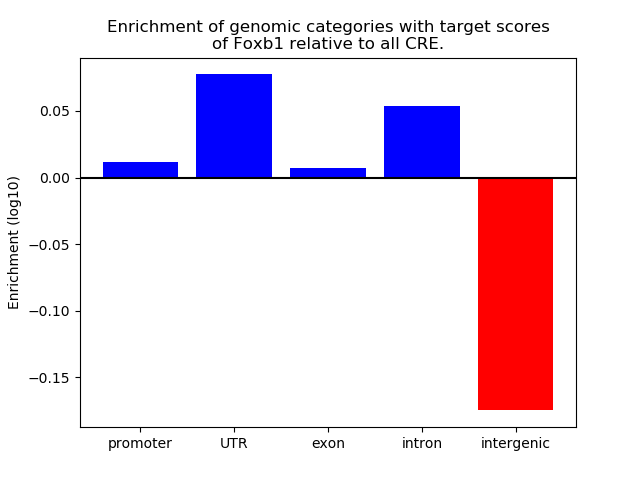
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_6881874_6882908 | 3.05 |
Gm13389 |
predicted gene 13389 |
1879 |
0.3 |
| chr13_83734808_83735118 | 2.50 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2397 |
0.18 |
| chr1_66388178_66389004 | 2.42 |
Map2 |
microtubule-associated protein 2 |
1580 |
0.42 |
| chr10_92164315_92164805 | 2.24 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
106 |
0.97 |
| chr4_6986587_6987701 | 2.07 |
Tox |
thymocyte selection-associated high mobility group box |
3339 |
0.35 |
| chr3_63897818_63898430 | 2.03 |
Plch1 |
phospholipase C, eta 1 |
1249 |
0.38 |
| chr13_109442519_109443753 | 1.84 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
953 |
0.73 |
| chr11_24087050_24087949 | 1.78 |
Bcl11a |
B cell CLL/lymphoma 11A (zinc finger protein) |
6829 |
0.15 |
| chr13_78192326_78192477 | 1.78 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
1958 |
0.24 |
| chr2_136052810_136053490 | 1.77 |
Lamp5 |
lysosomal-associated membrane protein family, member 5 |
911 |
0.62 |
| chr13_98356794_98358725 | 1.75 |
Foxd1 |
forkhead box D1 |
3517 |
0.19 |
| chr2_136713069_136714459 | 1.73 |
Snap25 |
synaptosomal-associated protein 25 |
286 |
0.92 |
| chrX_136591171_136591322 | 1.72 |
Tceal3 |
transcription elongation factor A (SII)-like 3 |
391 |
0.77 |
| chr3_13946382_13947629 | 1.60 |
Ralyl |
RALY RNA binding protein-like |
594 |
0.84 |
| chr1_77511833_77511984 | 1.57 |
Epha4 |
Eph receptor A4 |
3171 |
0.23 |
| chrX_166346283_166346827 | 1.57 |
Gpm6b |
glycoprotein m6b |
1713 |
0.43 |
| chr9_36821403_36822795 | 1.56 |
Fez1 |
fasciculation and elongation protein zeta 1 (zygin I) |
235 |
0.9 |
| chr18_57262576_57262848 | 1.54 |
Gm50200 |
predicted gene, 50200 |
52428 |
0.11 |
| chr3_34563437_34564219 | 1.50 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
3436 |
0.19 |
| chr4_22833789_22833940 | 1.47 |
Gm24078 |
predicted gene, 24078 |
86733 |
0.09 |
| chr19_4758474_4759201 | 1.25 |
Rbm4b |
RNA binding motif protein 4B |
1932 |
0.18 |
| chr11_120496309_120496677 | 1.25 |
Slc25a10 |
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
394 |
0.63 |
| chr14_21251974_21252125 | 1.23 |
Adk |
adenosine kinase |
66076 |
0.12 |
| chr11_76388616_76388907 | 1.22 |
Nxn |
nucleoredoxin |
10307 |
0.18 |
| chr2_65847409_65848267 | 1.17 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
1983 |
0.36 |
| chr7_79510881_79511376 | 1.16 |
A330074H02Rik |
RIKEN cDNA A330074H02 gene |
766 |
0.4 |
| chr14_122227888_122228085 | 1.15 |
1700108J01Rik |
RIKEN cDNA 1700108J01 gene |
5668 |
0.21 |
| chr13_34125172_34126139 | 1.14 |
Tubb2b |
tubulin, beta 2B class IIB |
4699 |
0.12 |
| chr6_136167149_136168437 | 1.13 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
4096 |
0.25 |
| chr13_84061710_84062414 | 1.11 |
Gm17750 |
predicted gene, 17750 |
2710 |
0.31 |
| chr3_4796861_4798079 | 1.10 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
88 |
0.98 |
| chr8_111539116_111539902 | 1.10 |
Znrf1 |
zinc and ring finger 1 |
1044 |
0.52 |
| chr3_118432535_118432944 | 1.09 |
Gm26871 |
predicted gene, 26871 |
1058 |
0.3 |
| chr16_74408614_74409150 | 1.07 |
Robo2 |
roundabout guidance receptor 2 |
2030 |
0.43 |
| chr3_68573207_68574269 | 1.07 |
Schip1 |
schwannomin interacting protein 1 |
1493 |
0.45 |
| chr13_94879818_94880134 | 1.05 |
Otp |
orthopedia homeobox |
2624 |
0.29 |
| chr1_138499487_138499975 | 1.04 |
Gm28501 |
predicted gene 28501 |
15884 |
0.2 |
| chr6_138420211_138420823 | 1.03 |
Lmo3 |
LIM domain only 3 |
935 |
0.55 |
| chr15_85675143_85675686 | 1.03 |
Lncppara |
long noncoding RNA near Ppara |
21798 |
0.12 |
| chr16_67617078_67617553 | 1.02 |
Cadm2 |
cell adhesion molecule 2 |
3178 |
0.32 |
| chr7_60390618_60391247 | 1.00 |
Gm30196 |
predicted gene, 30196 |
97728 |
0.06 |
| chr7_61309776_61310205 | 1.00 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
1723 |
0.5 |
| chr11_22284369_22284520 | 0.98 |
Ehbp1 |
EH domain binding protein 1 |
1394 |
0.55 |
| chr2_65565600_65566271 | 0.98 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
1557 |
0.45 |
| chr11_97841566_97842003 | 0.96 |
B230217C12Rik |
RIKEN cDNA B230217C12 gene |
465 |
0.67 |
| chr16_44429617_44430056 | 0.96 |
Cfap44 |
cilia and flagella associated protein 44 |
2207 |
0.36 |
| chr1_188186627_188186847 | 0.96 |
Gm38315 |
predicted gene, 38315 |
26030 |
0.21 |
| chr19_41850677_41851437 | 0.94 |
Frat2 |
frequently rearranged in advanced T cell lymphomas 2 |
2925 |
0.19 |
| chr10_19356598_19357962 | 0.94 |
Olig3 |
oligodendrocyte transcription factor 3 |
747 |
0.72 |
| chr4_138457222_138457745 | 0.93 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
3169 |
0.19 |
| chr2_48424016_48424809 | 0.91 |
Gm13481 |
predicted gene 13481 |
32833 |
0.17 |
| chr3_107041271_107042100 | 0.90 |
AI504432 |
expressed sequence AI504432 |
2181 |
0.26 |
| chr7_97842700_97844158 | 0.90 |
Pak1 |
p21 (RAC1) activated kinase 1 |
494 |
0.83 |
| chr4_62206894_62207045 | 0.88 |
Zfp37 |
zinc finger protein 37 |
920 |
0.55 |
| chr5_127347601_127347752 | 0.87 |
9430087B13Rik |
RIKEN cDNA 9430087B13 gene |
38659 |
0.17 |
| chr3_5223526_5223955 | 0.87 |
Zfhx4 |
zinc finger homeodomain 4 |
2235 |
0.28 |
| chr5_9723538_9723989 | 0.87 |
Grm3 |
glutamate receptor, metabotropic 3 |
1407 |
0.48 |
| chr9_62809372_62809735 | 0.86 |
Fem1b |
fem 1 homolog b |
2381 |
0.22 |
| chr8_108716860_108718878 | 0.85 |
Zfhx3 |
zinc finger homeobox 3 |
3225 |
0.3 |
| chr14_124191277_124191428 | 0.84 |
Fgf14 |
fibroblast growth factor 14 |
1550 |
0.55 |
| chr13_84060966_84061181 | 0.84 |
Gm17750 |
predicted gene, 17750 |
3699 |
0.27 |
| chr5_120429598_120430306 | 0.83 |
Lhx5 |
LIM homeobox protein 5 |
1747 |
0.23 |
| chr13_78189592_78191761 | 0.83 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
233 |
0.9 |
| chr2_174292352_174293472 | 0.82 |
Gnasas1 |
GNAS antisense RNA 1 |
2477 |
0.2 |
| chr15_54935690_54935949 | 0.81 |
Enpp2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
13206 |
0.17 |
| chr5_128603080_128603231 | 0.80 |
Fzd10 |
frizzled class receptor 10 |
2311 |
0.22 |
| chr18_31445651_31446131 | 0.80 |
Syt4 |
synaptotagmin IV |
1515 |
0.34 |
| chr12_117157079_117158175 | 0.79 |
Gm10421 |
predicted gene 10421 |
5976 |
0.31 |
| chr6_14904950_14905101 | 0.79 |
Foxp2 |
forkhead box P2 |
1568 |
0.56 |
| chr8_125997145_125997778 | 0.78 |
Kcnk1 |
potassium channel, subfamily K, member 1 |
2025 |
0.35 |
| chr16_4211315_4211466 | 0.78 |
Crebbp |
CREB binding protein |
2023 |
0.29 |
| chr4_22484307_22484937 | 0.78 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
3744 |
0.2 |
| chr3_7505462_7505760 | 0.78 |
Zc2hc1a |
zinc finger, C2HC-type containing 1A |
2125 |
0.29 |
| chr1_42691569_42692627 | 0.77 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
995 |
0.42 |
| chr16_77598670_77599917 | 0.77 |
Mir99a |
microRNA 99a |
357 |
0.44 |
| chr4_149589036_149589326 | 0.76 |
Clstn1 |
calsyntenin 1 |
1933 |
0.24 |
| chr14_75962509_75963193 | 0.76 |
Kctd4 |
potassium channel tetramerisation domain containing 4 |
7842 |
0.18 |
| chr7_127255013_127255164 | 0.76 |
Dctpp1 |
dCTP pyrophosphatase 1 |
5621 |
0.07 |
| chr5_17571826_17572147 | 0.76 |
Sema3c |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
2295 |
0.4 |
| chr18_69347722_69348433 | 0.75 |
Tcf4 |
transcription factor 4 |
179 |
0.97 |
| chr14_76420544_76421824 | 0.75 |
Tsc22d1 |
TSC22 domain family, member 1 |
2356 |
0.39 |
| chr17_35909041_35910037 | 0.75 |
Atat1 |
alpha tubulin acetyltransferase 1 |
34 |
0.91 |
| chr1_176355310_176355775 | 0.75 |
Gm17965 |
predicted gene, 17965 |
15899 |
0.19 |
| chr3_17791717_17791961 | 0.74 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
1882 |
0.32 |
| chr2_60122679_60122962 | 0.73 |
Gm13620 |
predicted gene 13620 |
2276 |
0.22 |
| chr17_48999728_48999879 | 0.73 |
Lrfn2 |
leucine rich repeat and fibronectin type III domain containing 2 |
67424 |
0.11 |
| chr19_32761292_32761490 | 0.73 |
Pten |
phosphatase and tensin homolog |
3796 |
0.28 |
| chr1_19107068_19107219 | 0.73 |
Tfap2d |
transcription factor AP-2, delta |
38 |
0.97 |
| chr1_81577415_81578042 | 0.73 |
Gm6198 |
predicted gene 6198 |
20245 |
0.25 |
| chr11_78694313_78696159 | 0.72 |
Nlk |
nemo like kinase |
1258 |
0.39 |
| chr2_74713881_74716227 | 0.72 |
Hoxd3os1 |
homeobox D3, opposite strand 1 |
942 |
0.23 |
| chr1_72536044_72537425 | 0.72 |
Marchf4 |
membrane associated ring-CH-type finger 4 |
196 |
0.95 |
| chr13_84058145_84058613 | 0.71 |
Gm17750 |
predicted gene, 17750 |
6393 |
0.23 |
| chr11_50175815_50175966 | 0.70 |
Mrnip |
MRN complex interacting protein |
983 |
0.4 |
| chr7_73554401_73554784 | 0.70 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
1328 |
0.3 |
| chrX_114476216_114476367 | 0.70 |
Klhl4 |
kelch-like 4 |
1687 |
0.43 |
| chr13_63557270_63560459 | 0.70 |
Ptch1 |
patched 1 |
4951 |
0.16 |
| chr5_131614206_131615000 | 0.68 |
2810432F15Rik |
RIKEN cDNA 2810432F15 gene |
835 |
0.43 |
| chr1_77495815_77496419 | 0.68 |
Mir6352 |
microRNA 6352 |
651 |
0.72 |
| chr7_62422629_62423664 | 0.68 |
Gm32061 |
predicted gene, 32061 |
99 |
0.96 |
| chr8_108706513_108707597 | 0.68 |
Zfhx3 |
zinc finger homeobox 3 |
3955 |
0.29 |
| chr7_60491777_60492274 | 0.68 |
Gm3198 |
predicted gene 3198 |
151866 |
0.03 |
| chr4_13752540_13753163 | 0.67 |
Runx1t1 |
RUNX1 translocation partner 1 |
1554 |
0.54 |
| chr9_50554623_50555585 | 0.67 |
Bco2 |
beta-carotene oxygenase 2 |
77 |
0.74 |
| chr5_33823212_33823938 | 0.66 |
Nsd2 |
nuclear receptor binding SET domain protein 2 |
2746 |
0.21 |
| chr2_45103994_45106973 | 0.65 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
4593 |
0.23 |
| chr3_68572046_68573169 | 0.65 |
Schip1 |
schwannomin interacting protein 1 |
362 |
0.89 |
| chr9_107229908_107230455 | 0.65 |
Dock3 |
dedicator of cyto-kinesis 3 |
1401 |
0.23 |
| chr8_109360134_109360931 | 0.64 |
Gm1943 |
predicted gene 1943 |
19668 |
0.22 |
| chr1_136343056_136343207 | 0.64 |
Camsap2 |
calmodulin regulated spectrin-associated protein family, member 2 |
2567 |
0.25 |
| chr18_69519778_69520273 | 0.64 |
Tcf4 |
transcription factor 4 |
204 |
0.96 |
| chrX_43429923_43430939 | 0.63 |
Tenm1 |
teneurin transmembrane protein 1 |
1305 |
0.49 |
| chr9_92174525_92174883 | 0.63 |
Plscr5 |
phospholipid scramblase family, member 5 |
18232 |
0.21 |
| chr13_83736071_83736534 | 0.63 |
Gm33366 |
predicted gene, 33366 |
2233 |
0.18 |
| chr14_60381648_60381975 | 0.63 |
Amer2 |
APC membrane recruitment 2 |
3525 |
0.27 |
| chr2_116053626_116054503 | 0.63 |
Meis2 |
Meis homeobox 2 |
4384 |
0.2 |
| chr3_33089775_33090447 | 0.63 |
Pex5l |
peroxisomal biogenesis factor 5-like |
7051 |
0.18 |
| chr5_93096288_93097671 | 0.62 |
Septin11 |
septin 11 |
3408 |
0.15 |
| chr12_44438829_44439626 | 0.62 |
Nrcam |
neuronal cell adhesion molecule |
152 |
0.96 |
| chr8_61309431_61310318 | 0.62 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
21002 |
0.18 |
| chrX_6169933_6171264 | 0.62 |
Nudt10 |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
2417 |
0.31 |
| chr7_78576803_78577063 | 0.62 |
Ntrk3 |
neurotrophic tyrosine kinase, receptor, type 3 |
850 |
0.49 |
| chr13_109445560_109445945 | 0.62 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
3569 |
0.38 |
| chr7_60938135_60938329 | 0.61 |
Gm44644 |
predicted gene 44644 |
15322 |
0.23 |
| chr7_144898015_144898977 | 0.60 |
Gm26793 |
predicted gene, 26793 |
1035 |
0.34 |
| chr2_118257147_118257384 | 0.60 |
Fsip1 |
fibrous sheath-interacting protein 1 |
299 |
0.86 |
| chr10_92162169_92163486 | 0.60 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
66 |
0.98 |
| chr13_83727321_83728283 | 0.60 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
304 |
0.83 |
| chr13_78205305_78205829 | 0.60 |
A830082K12Rik |
RIKEN cDNA A830082K12 gene |
2780 |
0.19 |
| chr19_12495109_12495388 | 0.60 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
6206 |
0.11 |
| chr13_56895147_56895774 | 0.60 |
Trpc7 |
transient receptor potential cation channel, subfamily C, member 7 |
277 |
0.94 |
| chr4_25797148_25797299 | 0.60 |
Fut9 |
fucosyltransferase 9 |
2632 |
0.27 |
| chr7_121705512_121705663 | 0.59 |
1700069B07Rik |
RIKEN cDNA 1700069B07 gene |
1488 |
0.32 |
| chr5_138618566_138618717 | 0.59 |
Gm42815 |
predicted gene 42815 |
647 |
0.47 |
| chr8_109250241_109250821 | 0.59 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
665 |
0.8 |
| chr5_84413029_84413589 | 0.59 |
Epha5 |
Eph receptor A5 |
3497 |
0.31 |
| chr19_54048025_54048767 | 0.58 |
Adra2a |
adrenergic receptor, alpha 2a |
3136 |
0.23 |
| chr18_23806890_23807163 | 0.58 |
Mapre2 |
microtubule-associated protein, RP/EB family, member 2 |
2923 |
0.23 |
| chr1_99775234_99775421 | 0.58 |
Cntnap5b |
contactin associated protein-like 5B |
2562 |
0.33 |
| chr19_12600891_12601046 | 0.58 |
Gm4952 |
predicted gene 4952 |
952 |
0.37 |
| chr4_22497544_22499061 | 0.58 |
Gm30731 |
predicted gene, 30731 |
7754 |
0.16 |
| chr4_114905731_114906737 | 0.58 |
Foxd2 |
forkhead box D2 |
2639 |
0.18 |
| chr6_47592948_47593162 | 0.58 |
Ezh2 |
enhancer of zeste 2 polycomb repressive complex 2 subunit |
1975 |
0.31 |
| chr4_44701489_44702654 | 0.57 |
5730488B01Rik |
RIKEN cDNA 5730488B01 gene |
729 |
0.51 |
| chr8_90957042_90957647 | 0.57 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
1909 |
0.31 |
| chr9_66351879_66352030 | 0.57 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
1428 |
0.42 |
| chr10_128015835_128016219 | 0.57 |
Prim1 |
DNA primase, p49 subunit |
831 |
0.38 |
| chr4_127027608_127028289 | 0.56 |
Sfpq |
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
1503 |
0.26 |
| chr5_131531462_131532675 | 0.56 |
Auts2 |
autism susceptibility candidate 2 |
2329 |
0.29 |
| chr9_41155678_41155975 | 0.56 |
Ubash3b |
ubiquitin associated and SH3 domain containing, B |
1676 |
0.27 |
| chr5_139548537_139550176 | 0.56 |
Uncx |
UNC homeobox |
5458 |
0.19 |
| chr5_122371006_122371157 | 0.56 |
Gpn3 |
GPN-loop GTPase 3 |
795 |
0.37 |
| chr2_92188323_92189316 | 0.56 |
Phf21a |
PHD finger protein 21A |
2354 |
0.24 |
| chr9_16485178_16485648 | 0.56 |
Fat3 |
FAT atypical cadherin 3 |
15872 |
0.27 |
| chr1_42707054_42709031 | 0.55 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
10 |
0.97 |
| chrX_101300806_101301090 | 0.55 |
Nlgn3 |
neuroligin 3 |
474 |
0.67 |
| chr14_16733494_16733722 | 0.55 |
Rarb |
retinoic acid receptor, beta |
85397 |
0.1 |
| chr3_87620769_87620920 | 0.55 |
Arhgef11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
2083 |
0.26 |
| chr14_76422028_76422179 | 0.54 |
Tsc22d1 |
TSC22 domain family, member 1 |
3275 |
0.34 |
| chr19_46570976_46571127 | 0.54 |
Arl3 |
ADP-ribosylation factor-like 3 |
2025 |
0.23 |
| chr3_146345424_146346260 | 0.54 |
Gm43334 |
predicted gene 43334 |
21422 |
0.13 |
| chr1_168676421_168677250 | 0.54 |
1700063I16Rik |
RIKEN cDNA 1700063I16 gene |
953 |
0.73 |
| chr4_154952453_154952604 | 0.54 |
Hes5 |
hes family bHLH transcription factor 5 |
8395 |
0.11 |
| chr8_26438640_26439210 | 0.53 |
Gm45580 |
predicted gene 45580 |
96 |
0.96 |
| chr9_71891771_71892231 | 0.53 |
Tcf12 |
transcription factor 12 |
3984 |
0.14 |
| chr12_118296552_118297427 | 0.53 |
Sp4 |
trans-acting transcription factor 4 |
4379 |
0.3 |
| chr1_42352204_42352355 | 0.53 |
Gm28140 |
predicted gene 28140 |
3658 |
0.31 |
| chr3_58576700_58577921 | 0.52 |
Selenot |
selenoprotein T |
612 |
0.68 |
| chr9_91372158_91372309 | 0.52 |
Zic4 |
zinc finger protein of the cerebellum 4 |
883 |
0.44 |
| chr9_66352035_66352703 | 0.52 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
1843 |
0.35 |
| chr17_58933106_58933374 | 0.52 |
Pdzph1 |
PDZ and pleckstrin homology domains 1 |
39745 |
0.16 |
| chr11_95631546_95631755 | 0.51 |
Gm11528 |
predicted gene 11528 |
8990 |
0.15 |
| chr8_108716106_108716847 | 0.51 |
Zfhx3 |
zinc finger homeobox 3 |
1832 |
0.41 |
| chr2_121268286_121268726 | 0.51 |
Trp53bp1 |
transformation related protein 53 binding protein 1 |
1780 |
0.25 |
| chr10_80208778_80209392 | 0.51 |
Pwwp3a |
PWWP domain containing 3A, DNA repair factor |
17349 |
0.07 |
| chr2_57125981_57126263 | 0.50 |
Nr4a2 |
nuclear receptor subfamily 4, group A, member 2 |
2119 |
0.32 |
| chr11_112708652_112709267 | 0.50 |
BC006965 |
cDNA sequence BC006965 |
1963 |
0.44 |
| chr8_111740274_111741466 | 0.50 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
2939 |
0.25 |
| chr9_35427115_35427995 | 0.50 |
Cdon |
cell adhesion molecule-related/down-regulated by oncogenes |
3967 |
0.19 |
| chr6_136170568_136170996 | 0.49 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
1107 |
0.5 |
| chr15_76076310_76077009 | 0.49 |
Puf60 |
poly-U binding splicing factor 60 |
294 |
0.76 |
| chr16_42338813_42338964 | 0.49 |
Gap43 |
growth associated protein 43 |
1763 |
0.45 |
| chr1_177449667_177450314 | 0.49 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
4169 |
0.18 |
| chr3_19310061_19310308 | 0.49 |
Pde7a |
phosphodiesterase 7A |
719 |
0.76 |
| chr3_31097203_31098928 | 0.48 |
Skil |
SKI-like |
1232 |
0.48 |
| chr15_60517604_60517755 | 0.48 |
Gm6392 |
predicted gene 6392 |
49555 |
0.15 |
| chr4_70530072_70530479 | 0.48 |
Megf9 |
multiple EGF-like-domains 9 |
4653 |
0.35 |
| chr11_69835212_69835923 | 0.48 |
Nlgn2 |
neuroligin 2 |
718 |
0.33 |
| chr2_155777177_155777352 | 0.48 |
Mmp24 |
matrix metallopeptidase 24 |
1922 |
0.21 |
| chr13_21928237_21928533 | 0.48 |
Gm44456 |
predicted gene, 44456 |
3369 |
0.09 |
| chr3_89689186_89689450 | 0.47 |
Adar |
adenosine deaminase, RNA-specific |
25704 |
0.13 |
| chr19_47016270_47016724 | 0.47 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
1344 |
0.28 |
| chr17_85394123_85395241 | 0.47 |
Rpl31-ps16 |
ribosomal protein L31, pseudogene 16 |
103951 |
0.07 |
| chr11_120576546_120578242 | 0.47 |
Arhgdia |
Rho GDP dissociation inhibitor (GDI) alpha |
3986 |
0.07 |
| chr1_66321652_66322390 | 0.46 |
Map2 |
microtubule-associated protein 2 |
23 |
0.98 |
| chr17_35920967_35921587 | 0.46 |
Mir1894 |
microRNA 1894 |
3388 |
0.07 |
| chr12_29529268_29529419 | 0.46 |
Myt1l |
myelin transcription factor 1-like |
942 |
0.56 |
| chr8_68059746_68060082 | 0.46 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
2313 |
0.38 |
| chr6_86031081_86032013 | 0.46 |
Add2 |
adducin 2 (beta) |
2800 |
0.16 |
| chr9_91368789_91369019 | 0.46 |
Zic4 |
zinc finger protein of the cerebellum 4 |
10 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.3 | 0.9 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.3 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 0.8 | GO:0015744 | succinate transport(GO:0015744) |
| 0.2 | 1.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.6 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 0.7 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 1.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.7 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 1.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.5 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 1.5 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 1.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.1 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.7 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.2 | GO:0072069 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.6 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.2 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.5 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.2 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.0 | GO:1902804 | negative regulation of synaptic vesicle transport(GO:1902804) |
| 0.0 | 0.0 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.2 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:2000978 | negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.4 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0006227 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.0 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.0 | GO:0003140 | determination of left/right asymmetry in lateral mesoderm(GO:0003140) nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.0 | GO:0043307 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 1.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 2.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 0.8 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.2 | 1.8 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 1.0 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 1.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 0.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0034874 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.8 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0043888 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |