Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
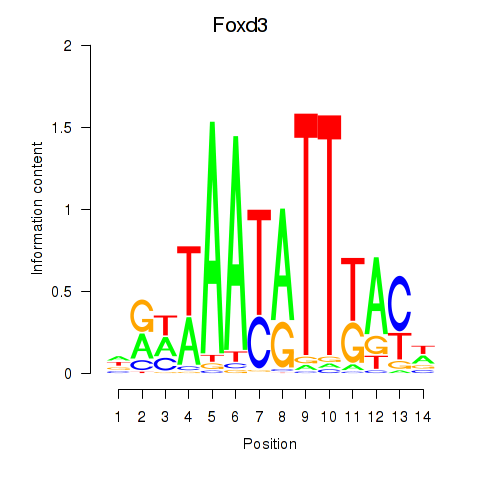
Results for Foxd3
Z-value: 0.54
Transcription factors associated with Foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd3
|
ENSMUSG00000067261.3 | forkhead box D3 |
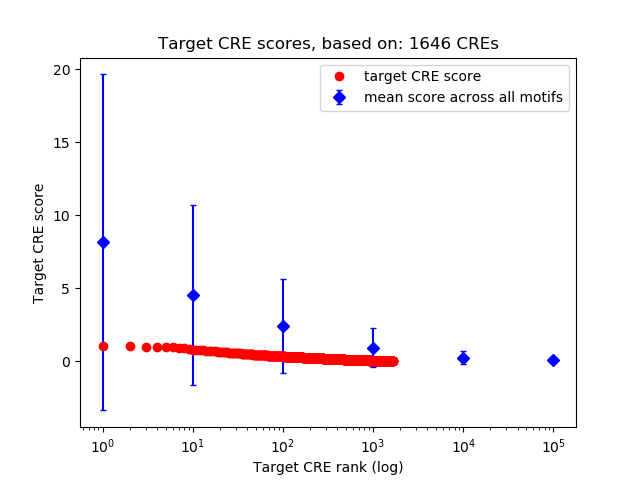
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_99655774_99658132 | Foxd3 | 654 | 0.428084 | 0.44 | 4.8e-04 | Click! |
Activity of the Foxd3 motif across conditions
Conditions sorted by the z-value of the Foxd3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
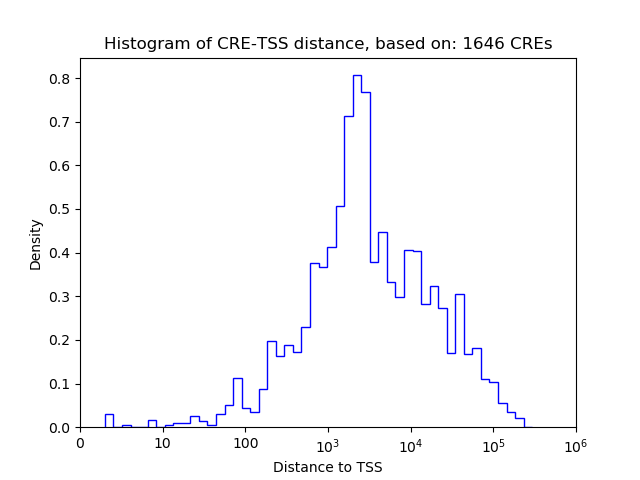
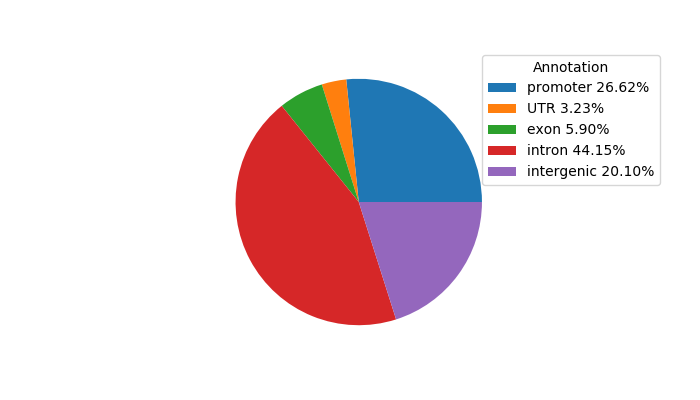
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_59095360_59096062 | 1.08 |
Mia2 |
MIA SH3 domain ER export factor 2 |
88 |
0.93 |
| chr6_37645617_37646179 | 1.02 |
Ybx1-ps2 |
Y box protein 1, pseudogene 2 |
41297 |
0.18 |
| chr14_110753664_110754027 | 1.00 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
1304 |
0.42 |
| chr11_96316025_96316970 | 0.99 |
Mir10a |
microRNA 10a |
668 |
0.4 |
| chr17_75180734_75181322 | 0.97 |
Ltbp1 |
latent transforming growth factor beta binding protein 1 |
2117 |
0.38 |
| chr2_74748525_74748913 | 0.95 |
Haglr |
Hoxd antisense growth associated long non-coding RNA |
1787 |
0.13 |
| chr8_32004165_32004671 | 0.89 |
Nrg1 |
neuregulin 1 |
4688 |
0.33 |
| chr6_137570111_137570964 | 0.89 |
Eps8 |
epidermal growth factor receptor pathway substrate 8 |
222 |
0.95 |
| chr13_98691032_98691862 | 0.84 |
Tmem171 |
transmembrane protein 171 |
3321 |
0.18 |
| chr13_17691689_17691840 | 0.79 |
Sugct |
succinyl-CoA glutarate-CoA transferase |
2972 |
0.18 |
| chr18_11049995_11051717 | 0.79 |
Gata6os |
GATA binding protein 6, opposite strand |
631 |
0.64 |
| chr12_31267815_31268221 | 0.77 |
Lamb1 |
laminin B1 |
2721 |
0.18 |
| chr7_89405888_89406433 | 0.76 |
Fzd4 |
frizzled class receptor 4 |
1805 |
0.24 |
| chr10_5287995_5289689 | 0.72 |
Gm23573 |
predicted gene, 23573 |
68331 |
0.12 |
| chr18_43736152_43737245 | 0.69 |
Spink1 |
serine peptidase inhibitor, Kazal type 1 |
859 |
0.6 |
| chr16_22160432_22161001 | 0.68 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
734 |
0.66 |
| chr5_101700539_101700765 | 0.68 |
9430085M18Rik |
RIKEN cDNA 9430085M18 gene |
2934 |
0.22 |
| chr7_36694259_36694943 | 0.68 |
Gm38285 |
predicted gene, 38285 |
837 |
0.52 |
| chr16_70798756_70798936 | 0.67 |
Gm49665 |
predicted gene, 49665 |
51124 |
0.17 |
| chr7_131032170_131032380 | 0.66 |
Dmbt1 |
deleted in malignant brain tumors 1 |
206 |
0.95 |
| chr10_69321361_69322099 | 0.64 |
Gm40685 |
predicted gene, 40685 |
9591 |
0.17 |
| chr13_55923232_55923960 | 0.62 |
Gm47071 |
predicted gene, 47071 |
78669 |
0.07 |
| chr14_74913536_74914053 | 0.61 |
Gm10847 |
predicted gene 10847 |
12651 |
0.19 |
| chr10_116276295_116276851 | 0.61 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
1023 |
0.54 |
| chr17_75437948_75438432 | 0.60 |
Rasgrp3 |
RAS, guanyl releasing protein 3 |
2264 |
0.4 |
| chr4_53878509_53878668 | 0.58 |
Gm12468 |
predicted gene 12468 |
663 |
0.67 |
| chr14_110753127_110753447 | 0.58 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
1862 |
0.32 |
| chr1_184729275_184729496 | 0.57 |
Hlx |
H2.0-like homeobox |
2213 |
0.23 |
| chr8_121129194_121129579 | 0.56 |
Foxl1 |
forkhead box L1 |
1446 |
0.3 |
| chr12_57544414_57545119 | 0.56 |
Foxa1 |
forkhead box A1 |
1355 |
0.37 |
| chr6_137935280_137935834 | 0.55 |
Gm24308 |
predicted gene, 24308 |
15586 |
0.26 |
| chr15_103058659_103059955 | 0.55 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
1 |
0.95 |
| chr8_47763533_47764764 | 0.54 |
Gm45824 |
predicted gene 45824 |
1977 |
0.21 |
| chr9_60795900_60796901 | 0.54 |
Uaca |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
1775 |
0.35 |
| chr4_84768134_84768574 | 0.53 |
Gm12415 |
predicted gene 12415 |
26879 |
0.21 |
| chr10_116275477_116276105 | 0.52 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
241 |
0.93 |
| chr17_88628874_88630550 | 0.52 |
Ston1 |
stonin 1 |
2348 |
0.25 |
| chr15_51274333_51274786 | 0.51 |
Gm19303 |
predicted gene, 19303 |
121051 |
0.06 |
| chr8_33905601_33906425 | 0.50 |
Rbpms |
RNA binding protein gene with multiple splicing |
14249 |
0.16 |
| chr17_78205213_78205510 | 0.50 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
4296 |
0.17 |
| chr6_142570795_142570980 | 0.50 |
Kcnj8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
484 |
0.82 |
| chr17_79261126_79261277 | 0.49 |
Gm5230 |
predicted gene 5230 |
3152 |
0.32 |
| chr4_140003980_140005143 | 0.48 |
Klhdc7a |
kelch domain containing 7A |
36535 |
0.13 |
| chr2_156047784_156048442 | 0.47 |
Fer1l4 |
fer-1-like 4 (C. elegans) |
4834 |
0.1 |
| chr9_77044031_77044308 | 0.47 |
Tinag |
tubulointerstitial nephritis antigen |
1580 |
0.34 |
| chr7_143086048_143086508 | 0.47 |
Trpm5 |
transient receptor potential cation channel, subfamily M, member 5 |
851 |
0.44 |
| chr5_139598854_139600619 | 0.46 |
Uncx |
UNC homeobox |
55838 |
0.1 |
| chr13_48543356_48543507 | 0.46 |
Gm37238 |
predicted gene, 37238 |
1602 |
0.19 |
| chr16_34999592_34999743 | 0.46 |
Mylk |
myosin, light polypeptide kinase |
4860 |
0.18 |
| chr11_30195109_30195350 | 0.46 |
Sptbn1 |
spectrin beta, non-erythrocytic 1 |
3028 |
0.33 |
| chr6_100195944_100196095 | 0.45 |
Rybp |
RING1 and YY1 binding protein |
37112 |
0.14 |
| chr18_83984951_83985102 | 0.45 |
Gm50421 |
predicted gene, 50421 |
2969 |
0.31 |
| chr12_80123554_80123705 | 0.45 |
2310015A10Rik |
RIKEN cDNA 2310015A10 gene |
2490 |
0.19 |
| chr10_99618893_99619600 | 0.44 |
Gm20110 |
predicted gene, 20110 |
10075 |
0.17 |
| chr2_170139645_170139884 | 0.44 |
Zfp217 |
zinc finger protein 217 |
2914 |
0.38 |
| chr13_71951907_71952058 | 0.43 |
Gm40999 |
predicted gene, 40999 |
4570 |
0.27 |
| chr3_145101984_145102292 | 0.43 |
Clca2 |
chloride channel accessory 2 |
2695 |
0.28 |
| chr15_101139509_101140276 | 0.43 |
Acvrl1 |
activin A receptor, type II-like 1 |
2292 |
0.19 |
| chr2_125154199_125154495 | 0.43 |
Slc12a1 |
solute carrier family 12, member 1 |
1747 |
0.28 |
| chr12_37112722_37113009 | 0.42 |
Meox2 |
mesenchyme homeobox 2 |
4325 |
0.22 |
| chr2_170075358_170075771 | 0.42 |
Zfp217 |
zinc finger protein 217 |
55656 |
0.15 |
| chr19_46532996_46533147 | 0.42 |
Arl3 |
ADP-ribosylation factor-like 3 |
9742 |
0.14 |
| chr19_46570976_46571127 | 0.41 |
Arl3 |
ADP-ribosylation factor-like 3 |
2025 |
0.23 |
| chr1_155809619_155810012 | 0.41 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
3005 |
0.17 |
| chr10_97566192_97567945 | 0.41 |
Lum |
lumican |
1940 |
0.32 |
| chr5_28462243_28464057 | 0.41 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
3834 |
0.23 |
| chr9_77464536_77464687 | 0.40 |
Lrrc1 |
leucine rich repeat containing 1 |
22120 |
0.17 |
| chr1_171446655_171447030 | 0.40 |
F11r |
F11 receptor |
9263 |
0.09 |
| chr6_99275359_99276069 | 0.39 |
Foxp1 |
forkhead box P1 |
9182 |
0.29 |
| chr10_97479376_97480642 | 0.39 |
Dcn |
decorin |
400 |
0.88 |
| chr19_25381686_25382798 | 0.39 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
13061 |
0.21 |
| chr16_26103797_26103994 | 0.39 |
P3h2 |
prolyl 3-hydroxylase 2 |
1889 |
0.46 |
| chr5_47988429_47989444 | 0.39 |
Slit2 |
slit guidance ligand 2 |
1094 |
0.46 |
| chr13_89585655_89585816 | 0.39 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
1224 |
0.51 |
| chr18_61954762_61955556 | 0.39 |
Sh3tc2 |
SH3 domain and tetratricopeptide repeats 2 |
2075 |
0.33 |
| chr18_56649239_56649419 | 0.38 |
Gm50289 |
predicted gene, 50289 |
18449 |
0.14 |
| chr9_31688091_31688790 | 0.38 |
Gm47436 |
predicted gene, 47436 |
10414 |
0.21 |
| chr1_162220626_162221297 | 0.38 |
Dnm3os |
dynamin 3, opposite strand |
1093 |
0.42 |
| chr18_90002590_90002741 | 0.38 |
Gm6173 |
predicted gene 6173 |
16414 |
0.25 |
| chr10_44525081_44525285 | 0.38 |
Prdm1 |
PR domain containing 1, with ZNF domain |
3318 |
0.22 |
| chr11_96292706_96294113 | 0.38 |
Hoxb6 |
homeobox B6 |
933 |
0.29 |
| chr9_70241032_70241795 | 0.37 |
Myo1e |
myosin IE |
34045 |
0.17 |
| chr2_74707355_74707963 | 0.37 |
Hoxd8 |
homeobox D8 |
2507 |
0.08 |
| chr12_98683337_98683705 | 0.37 |
Ptpn21 |
protein tyrosine phosphatase, non-receptor type 21 |
229 |
0.9 |
| chr9_92694521_92694861 | 0.37 |
Gm29396 |
predicted gene 29396 |
2748 |
0.29 |
| chrX_146014065_146014216 | 0.37 |
Gm6447 |
predicted gene 6447 |
19849 |
0.18 |
| chrX_120591703_120591854 | 0.37 |
Gm14930 |
predicted gene 14930 |
149092 |
0.04 |
| chr12_35533032_35533561 | 0.37 |
Gm48236 |
predicted gene, 48236 |
1125 |
0.37 |
| chr10_5594267_5594984 | 0.37 |
Myct1 |
myc target 1 |
850 |
0.67 |
| chr6_52172116_52173481 | 0.36 |
Gm15050 |
predicted gene 15050 |
83 |
0.89 |
| chr6_99517992_99518143 | 0.36 |
Foxp1 |
forkhead box P1 |
2930 |
0.29 |
| chr13_111326182_111326588 | 0.36 |
Gm15285 |
predicted gene 15285 |
35290 |
0.13 |
| chr2_32908170_32908947 | 0.35 |
Fam129b |
family with sequence similarity 129, member B |
2350 |
0.19 |
| chr2_125282385_125283036 | 0.35 |
Gm14003 |
predicted gene 14003 |
1268 |
0.44 |
| chr2_36233437_36233701 | 0.35 |
Ptgs1 |
prostaglandin-endoperoxide synthase 1 |
2680 |
0.17 |
| chr15_83530746_83530897 | 0.34 |
Bik |
BCL2-interacting killer |
2723 |
0.16 |
| chr10_121982673_121982824 | 0.34 |
Srgap1 |
SLIT-ROBO Rho GTPase activating protein 1 |
64560 |
0.09 |
| chr5_139548537_139550176 | 0.34 |
Uncx |
UNC homeobox |
5458 |
0.19 |
| chr18_84077214_84077748 | 0.34 |
Tshz1 |
teashirt zinc finger family member 1 |
7594 |
0.17 |
| chrX_82950582_82950733 | 0.34 |
Dmd |
dystrophin, muscular dystrophy |
1754 |
0.53 |
| chr19_55102740_55102891 | 0.33 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
276 |
0.91 |
| chr15_82793644_82794188 | 0.33 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
306 |
0.81 |
| chr15_3580312_3581004 | 0.33 |
Ghr |
growth hormone receptor |
1184 |
0.57 |
| chr5_113799288_113800716 | 0.33 |
Tmem119 |
transmembrane protein 119 |
444 |
0.69 |
| chr6_141380917_141381972 | 0.33 |
Gm10400 |
predicted gene 10400 |
40891 |
0.17 |
| chr7_135526475_135527168 | 0.33 |
Clrn3 |
clarin 3 |
1833 |
0.31 |
| chr16_40048843_40048994 | 0.32 |
Lsamp |
limbic system-associated membrane protein |
64323 |
0.13 |
| chr12_110488357_110488534 | 0.32 |
Gm19605 |
predicted gene, 19605 |
2237 |
0.25 |
| chr9_43231799_43232208 | 0.32 |
Oaf |
out at first homolog |
6911 |
0.15 |
| chr2_68435653_68436705 | 0.32 |
Stk39 |
serine/threonine kinase 39 |
35761 |
0.16 |
| chr6_101374596_101375205 | 0.32 |
Pdzrn3 |
PDZ domain containing RING finger 3 |
2442 |
0.26 |
| chr5_24725084_24725454 | 0.32 |
Wdr86 |
WD repeat domain 86 |
5411 |
0.15 |
| chr9_61366071_61366222 | 0.32 |
Gm10655 |
predicted gene 10655 |
5481 |
0.2 |
| chr2_169741294_169741445 | 0.31 |
Tshz2 |
teashirt zinc finger family member 2 |
107693 |
0.07 |
| chr1_184675496_184676152 | 0.31 |
Gm38358 |
predicted gene, 38358 |
19210 |
0.14 |
| chr8_11309999_11311023 | 0.31 |
Col4a1 |
collagen, type IV, alpha 1 |
2178 |
0.24 |
| chr7_112498586_112498737 | 0.31 |
Parva |
parvin, alpha |
21040 |
0.2 |
| chr2_110360491_110361293 | 0.31 |
Fibin |
fin bud initiation factor homolog (zebrafish) |
2291 |
0.34 |
| chr4_153018259_153019185 | 0.31 |
Gm25779 |
predicted gene, 25779 |
5138 |
0.33 |
| chr15_84812976_84813127 | 0.31 |
Phf21b |
PHD finger protein 21B |
9494 |
0.17 |
| chr19_10699827_10700127 | 0.31 |
Vps37c |
vacuolar protein sorting 37C |
11212 |
0.1 |
| chr11_35544219_35545325 | 0.31 |
Slit3 |
slit guidance ligand 3 |
87 |
0.98 |
| chr2_160620140_160620291 | 0.30 |
Gm14221 |
predicted gene 14221 |
244 |
0.91 |
| chr15_8967409_8968940 | 0.30 |
Ranbp3l |
RAN binding protein 3-like |
172 |
0.96 |
| chr4_118583799_118583950 | 0.30 |
Cfap57 |
cilia and flagella associated protein 57 |
36531 |
0.08 |
| chr5_124204256_124204407 | 0.30 |
Pitpnm2os2 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 2 |
8689 |
0.12 |
| chr14_45511039_45511190 | 0.30 |
Fermt2 |
fermitin family member 2 |
7826 |
0.11 |
| chr6_4953094_4953370 | 0.29 |
Gm26556 |
predicted gene, 26556 |
5751 |
0.16 |
| chr11_58380480_58380717 | 0.29 |
Lypd8 |
LY6/PLAUR domain containing 8 |
1555 |
0.22 |
| chr7_120916926_120917434 | 0.29 |
Polr3e |
polymerase (RNA) III (DNA directed) polypeptide E |
564 |
0.66 |
| chr2_115856008_115856929 | 0.29 |
Meis2 |
Meis homeobox 2 |
12399 |
0.29 |
| chr5_147292277_147292672 | 0.29 |
Mir7k |
microRNA 7k |
9176 |
0.1 |
| chr6_52182052_52182511 | 0.29 |
Gm29430 |
predicted gene 29430 |
829 |
0.26 |
| chr6_129183729_129184848 | 0.29 |
Clec2d |
C-type lectin domain family 2, member d |
3673 |
0.14 |
| chr8_26329826_26330767 | 0.29 |
Gm31784 |
predicted gene, 31784 |
17962 |
0.15 |
| chr2_126556967_126557245 | 0.28 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
978 |
0.57 |
| chr6_13193034_13193185 | 0.28 |
Vwde |
von Willebrand factor D and EGF domains |
15434 |
0.23 |
| chr19_20602564_20602913 | 0.28 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
777 |
0.68 |
| chr8_108104785_108105045 | 0.28 |
Zfhx3 |
zinc finger homeobox 3 |
110356 |
0.07 |
| chr9_96885174_96885371 | 0.28 |
Pxylp1 |
2-phosphoxylose phosphatase 1 |
4141 |
0.18 |
| chr10_80330985_80331440 | 0.28 |
Reep6 |
receptor accessory protein 6 |
1010 |
0.24 |
| chr1_74964735_74965250 | 0.28 |
Gm37744 |
predicted gene, 37744 |
10740 |
0.12 |
| chr14_16572461_16573738 | 0.28 |
Rarb |
retinoic acid receptor, beta |
1946 |
0.34 |
| chr10_75930797_75931493 | 0.28 |
Mmp11 |
matrix metallopeptidase 11 |
1330 |
0.2 |
| chr4_118487482_118487969 | 0.28 |
Tie1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
2030 |
0.19 |
| chr4_45965494_45966277 | 0.27 |
Tdrd7 |
tudor domain containing 7 |
551 |
0.79 |
| chr14_44863488_44863639 | 0.27 |
Ptgdr |
prostaglandin D receptor |
4188 |
0.12 |
| chr6_100525882_100526106 | 0.27 |
1700049E22Rik |
RIKEN cDNA 1700049E22 gene |
1406 |
0.41 |
| chr16_14710279_14710697 | 0.27 |
Snai2 |
snail family zinc finger 2 |
4636 |
0.28 |
| chr12_39741189_39741358 | 0.27 |
Gm18116 |
predicted gene, 18116 |
23913 |
0.23 |
| chr8_106521568_106521719 | 0.27 |
Cdh3 |
cadherin 3 |
10752 |
0.17 |
| chr5_17642536_17642831 | 0.27 |
Sema3c |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
51519 |
0.17 |
| chr3_60529662_60530216 | 0.27 |
Mbnl1 |
muscleblind like splicing factor 1 |
308 |
0.91 |
| chr2_148043571_148045987 | 0.27 |
Foxa2 |
forkhead box A2 |
685 |
0.65 |
| chr9_97276072_97276223 | 0.27 |
A030012G06Rik |
RIKEN cDNA A030012G06 gene |
13039 |
0.17 |
| chr9_31911798_31912140 | 0.27 |
Barx2 |
BarH-like homeobox 2 |
1493 |
0.35 |
| chr9_114560970_114561936 | 0.27 |
Trim71 |
tripartite motif-containing 71 |
2916 |
0.23 |
| chr7_70369044_70370707 | 0.27 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
3140 |
0.16 |
| chr14_20699000_20699218 | 0.27 |
Gm25328 |
predicted gene, 25328 |
2414 |
0.11 |
| chr8_41015082_41015919 | 0.27 |
Mtus1 |
mitochondrial tumor suppressor 1 |
820 |
0.53 |
| chr2_166030364_166031781 | 0.26 |
Ncoa3 |
nuclear receptor coactivator 3 |
16846 |
0.15 |
| chr13_38126437_38126720 | 0.26 |
Dsp |
desmoplakin |
24716 |
0.14 |
| chr10_79531492_79531759 | 0.26 |
Mir6909 |
microRNA 6909 |
535 |
0.64 |
| chr9_58346541_58346692 | 0.26 |
Tbc1d21 |
TBC1 domain family, member 21 |
23753 |
0.13 |
| chr16_21818680_21818960 | 0.26 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
7122 |
0.12 |
| chr12_111911009_111911160 | 0.26 |
Ppp1r13b |
protein phosphatase 1, regulatory subunit 13B |
2974 |
0.16 |
| chr4_116144678_116144948 | 0.26 |
Lurap1 |
leucine rich adaptor protein 1 |
204 |
0.88 |
| chr3_146946599_146946750 | 0.26 |
Ttll7 |
tubulin tyrosine ligase-like family, member 7 |
2754 |
0.35 |
| chr18_54452409_54453689 | 0.26 |
Gm50361 |
predicted gene, 50361 |
23716 |
0.19 |
| chr7_12809561_12809753 | 0.26 |
Zfp329 |
zinc finger protein 329 |
1582 |
0.17 |
| chr4_8237536_8238080 | 0.26 |
Car8 |
carbonic anhydrase 8 |
1233 |
0.46 |
| chr15_38567059_38567529 | 0.25 |
Gm29697 |
predicted gene, 29697 |
6702 |
0.13 |
| chr5_92899429_92899666 | 0.25 |
Shroom3 |
shroom family member 3 |
1554 |
0.45 |
| chr17_67351548_67352402 | 0.25 |
Ptprm |
protein tyrosine phosphatase, receptor type, M |
2098 |
0.47 |
| chr1_153279868_153280137 | 0.25 |
Gm38009 |
predicted gene, 38009 |
7188 |
0.18 |
| chr2_103298575_103299104 | 0.25 |
Ehf |
ets homologous factor |
726 |
0.68 |
| chr7_46029818_46030744 | 0.25 |
Abcc6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
21 |
0.96 |
| chr12_75537153_75537454 | 0.25 |
Ppp2r5e |
protein phosphatase 2, regulatory subunit B', epsilon |
12371 |
0.19 |
| chr9_19125111_19125262 | 0.25 |
Olfr836 |
olfactory receptor 836 |
4229 |
0.16 |
| chr11_112708652_112709267 | 0.25 |
BC006965 |
cDNA sequence BC006965 |
1963 |
0.44 |
| chr3_24573795_24573946 | 0.25 |
Gm24704 |
predicted gene, 24704 |
11796 |
0.31 |
| chrX_122831261_122831412 | 0.25 |
Gm14940 |
predicted gene 14940 |
62467 |
0.15 |
| chr2_4588036_4588187 | 0.25 |
Gm13179 |
predicted gene 13179 |
824 |
0.64 |
| chr14_105744608_105744759 | 0.25 |
Gm10076 |
predicted gene 10076 |
62854 |
0.1 |
| chr16_87716187_87716338 | 0.25 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
17258 |
0.2 |
| chr4_83050103_83050875 | 0.24 |
Frem1 |
Fras1 related extracellular matrix protein 1 |
1678 |
0.41 |
| chr8_108701364_108702549 | 0.24 |
Zfhx3 |
zinc finger homeobox 3 |
1144 |
0.58 |
| chr3_123984120_123984365 | 0.24 |
Gm15399 |
predicted gene 15399 |
862 |
0.74 |
| chr13_25083128_25083551 | 0.24 |
Dcdc2a |
doublecortin domain containing 2a |
13008 |
0.18 |
| chr11_96321472_96321760 | 0.24 |
Hoxb3 |
homeobox B3 |
1710 |
0.14 |
| chr10_99912846_99913873 | 0.24 |
Gm47579 |
predicted gene, 47579 |
47339 |
0.13 |
| chrX_36587406_36587686 | 0.24 |
Pgrmc1 |
progesterone receptor membrane component 1 |
10660 |
0.2 |
| chr4_135254409_135255495 | 0.24 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
17862 |
0.14 |
| chr10_115817324_115818606 | 0.24 |
Tspan8 |
tetraspanin 8 |
681 |
0.78 |
| chr2_37742222_37742373 | 0.24 |
Crb2 |
crumbs family member 2 |
33952 |
0.13 |
| chr4_118691378_118691529 | 0.24 |
Olfr1342 |
olfactory receptor 1342 |
990 |
0.36 |
| chr5_124892161_124892312 | 0.24 |
Zfp664 |
zinc finger protein 664 |
7667 |
0.19 |
| chr16_4644396_4644547 | 0.24 |
Coro7 |
coronin 7 |
2245 |
0.18 |
| chr17_56277120_56277484 | 0.24 |
Ticam1 |
toll-like receptor adaptor molecule 1 |
516 |
0.61 |
| chr6_99095299_99096526 | 0.23 |
Foxp1 |
forkhead box P1 |
293 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0060459 | left lung development(GO:0060459) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.1 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.1 | GO:0032905 | transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 0.1 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0048619 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 1.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |