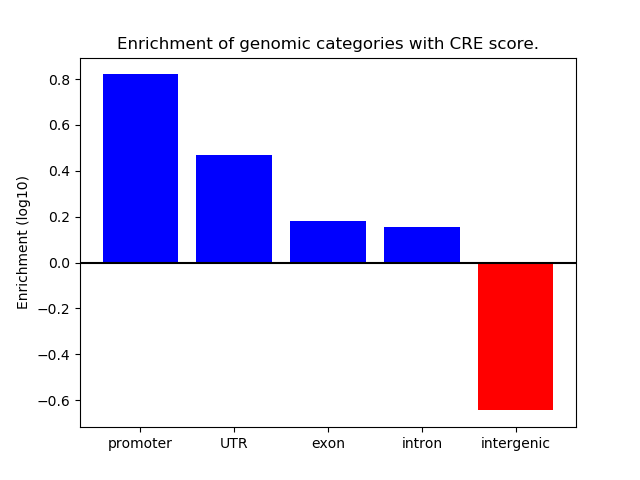
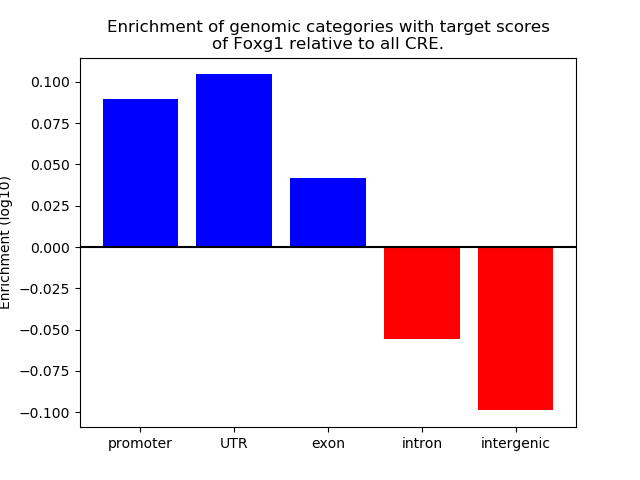
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
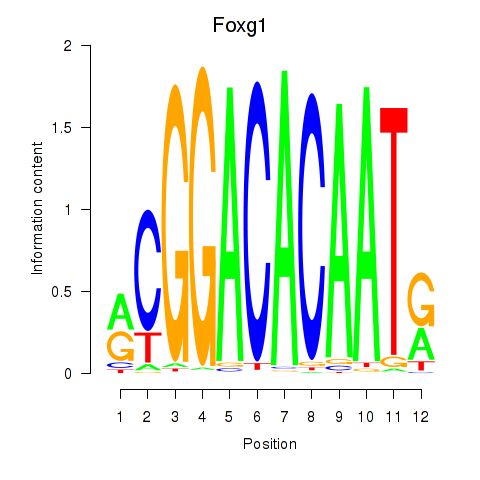
Results for Foxg1
Z-value: 0.67
Transcription factors associated with Foxg1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxg1
|
ENSMUSG00000020950.9 | forkhead box G1 |
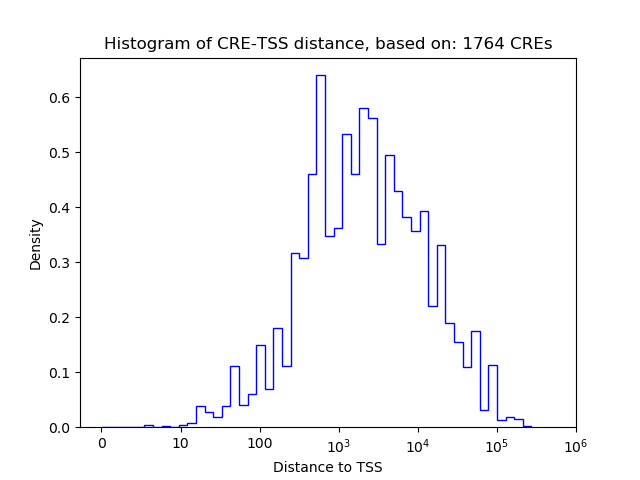
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_49382714_49384085 | Foxg1 | 392 | 0.722333 | -0.02 | 8.9e-01 | Click! |
Activity of the Foxg1 motif across conditions
Conditions sorted by the z-value of the Foxg1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
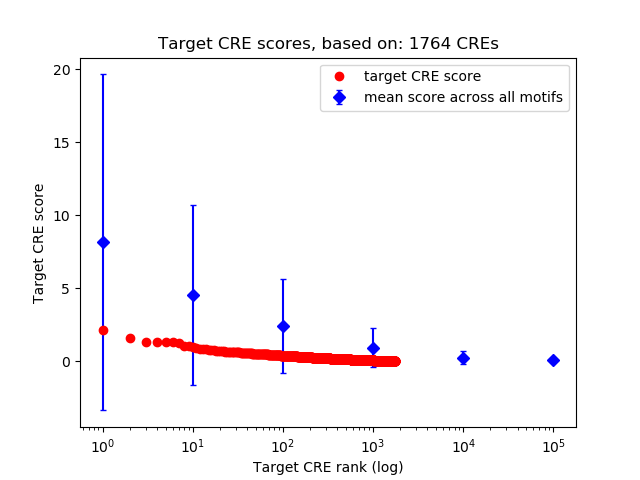
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_57784547_57786586 | 2.14 |
Fabp7 |
fatty acid binding protein 7, brain |
643 |
0.68 |
| chr16_18429039_18430122 | 1.61 |
Txnrd2 |
thioredoxin reductase 2 |
655 |
0.54 |
| chr8_80494890_80495139 | 1.35 |
Gypa |
glycophorin A |
1233 |
0.53 |
| chr7_78914724_78915335 | 1.33 |
Isg20 |
interferon-stimulated protein |
700 |
0.57 |
| chr8_80497324_80498362 | 1.31 |
Gypa |
glycophorin A |
4062 |
0.27 |
| chr6_118757029_118757180 | 1.29 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
870 |
0.71 |
| chr7_62422629_62423664 | 1.25 |
Gm32061 |
predicted gene, 32061 |
99 |
0.96 |
| chr1_168287219_168287370 | 1.04 |
Gm37524 |
predicted gene, 37524 |
50377 |
0.16 |
| chr11_102364387_102365146 | 1.03 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
481 |
0.67 |
| chr11_54026747_54027441 | 0.96 |
Slc22a4 |
solute carrier family 22 (organic cation transporter), member 4 |
858 |
0.54 |
| chr2_105680581_105683424 | 0.94 |
Pax6 |
paired box 6 |
290 |
0.89 |
| chr7_143004834_143005031 | 0.87 |
Tspan32 |
tetraspanin 32 |
114 |
0.94 |
| chr8_80868370_80868642 | 0.87 |
Gm31105 |
predicted gene, 31105 |
11434 |
0.18 |
| chr13_41236642_41237224 | 0.84 |
Gm32063 |
predicted gene, 32063 |
588 |
0.6 |
| chr7_90083908_90084239 | 0.79 |
Gm5341 |
predicted pseudogene 5341 |
23895 |
0.11 |
| chr6_87776091_87777171 | 0.78 |
Gm43904 |
predicted gene, 43904 |
488 |
0.55 |
| chr9_91378153_91379783 | 0.76 |
Zic4 |
zinc finger protein of the cerebellum 4 |
326 |
0.81 |
| chr6_128526086_128526400 | 0.72 |
Pzp |
PZP, alpha-2-macroglobulin like |
460 |
0.65 |
| chr17_24202438_24203465 | 0.71 |
Tbc1d24 |
TBC1 domain family, member 24 |
2531 |
0.12 |
| chr6_52162888_52164000 | 0.70 |
Hoxa2 |
homeobox A2 |
1387 |
0.15 |
| chr2_74713881_74716227 | 0.69 |
Hoxd3os1 |
homeobox D3, opposite strand 1 |
942 |
0.23 |
| chr10_12962620_12962953 | 0.68 |
Stx11 |
syntaxin 11 |
1472 |
0.41 |
| chr10_91967800_91968283 | 0.67 |
Gm31592 |
predicted gene, 31592 |
79212 |
0.1 |
| chr11_74572004_74572226 | 0.66 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
18009 |
0.18 |
| chr12_116407284_116407647 | 0.66 |
Ncapg2 |
non-SMC condensin II complex, subunit G2 |
2031 |
0.24 |
| chr9_70934045_70934653 | 0.65 |
Lipc |
lipase, hepatic |
264 |
0.92 |
| chr4_119183706_119183857 | 0.65 |
Ermap |
erythroblast membrane-associated protein |
4966 |
0.1 |
| chr17_71525336_71525487 | 0.64 |
Ndc80 |
NDC80 kinetochore complex component |
1379 |
0.24 |
| chr4_141076285_141076997 | 0.64 |
Necap2 |
NECAP endocytosis associated 2 |
1584 |
0.24 |
| chr13_47104855_47106138 | 0.63 |
Dek |
DEK oncogene (DNA binding) |
334 |
0.73 |
| chr4_143045346_143045497 | 0.62 |
6330411D24Rik |
RIKEN cDNA 6330411D24 gene |
28345 |
0.19 |
| chr10_80570596_80572042 | 0.61 |
Klf16 |
Kruppel-like factor 16 |
6002 |
0.08 |
| chr5_116021804_116022431 | 0.61 |
Prkab1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
471 |
0.73 |
| chr14_109511936_109512087 | 0.60 |
Gm18977 |
predicted gene, 18977 |
11088 |
0.28 |
| chr18_63727695_63728070 | 0.59 |
Wdr7 |
WD repeat domain 7 |
11182 |
0.16 |
| chr3_85991395_85991798 | 0.59 |
Prss48 |
protease, serine 48 |
10895 |
0.12 |
| chr11_97471430_97471581 | 0.58 |
Arhgap23 |
Rho GTPase activating protein 23 |
19375 |
0.13 |
| chr9_107301461_107301858 | 0.58 |
Gm17041 |
predicted gene 17041 |
179 |
0.88 |
| chr2_26349516_26349790 | 0.57 |
Dnlz |
DNL-type zinc finger |
2447 |
0.12 |
| chr4_129463918_129464422 | 0.57 |
Bsdc1 |
BSD domain containing 1 |
372 |
0.74 |
| chr15_77756117_77756516 | 0.56 |
Apol8 |
apolipoprotein L 8 |
666 |
0.54 |
| chr12_24645782_24646298 | 0.55 |
Klf11 |
Kruppel-like factor 11 |
5234 |
0.16 |
| chr10_80569864_80570541 | 0.55 |
Klf16 |
Kruppel-like factor 16 |
7119 |
0.08 |
| chr6_54851742_54852146 | 0.54 |
Znrf2 |
zinc and ring finger 2 |
34496 |
0.15 |
| chr16_20713558_20714737 | 0.54 |
Clcn2 |
chloride channel, voltage-sensitive 2 |
479 |
0.59 |
| chr19_36896833_36896984 | 0.53 |
Tnks2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
16335 |
0.17 |
| chr4_119189472_119189950 | 0.53 |
Ermap |
erythroblast membrane-associated protein |
240 |
0.84 |
| chr5_116573497_116573648 | 0.53 |
Gm42854 |
predicted gene 42854 |
77 |
0.97 |
| chr7_45238275_45239242 | 0.53 |
Cd37 |
CD37 antigen |
38 |
0.92 |
| chr7_99803412_99803631 | 0.53 |
F730035P03Rik |
RIKEN cDNA F730035P03 gene |
21982 |
0.1 |
| chr9_43746633_43747189 | 0.52 |
Gm30015 |
predicted gene, 30015 |
811 |
0.54 |
| chr11_27966865_27967016 | 0.52 |
Gm12080 |
predicted gene 12080 |
79095 |
0.11 |
| chr4_130006445_130007362 | 0.51 |
Adgrb2 |
adhesion G protein-coupled receptor B2 |
2973 |
0.19 |
| chr12_69783268_69783598 | 0.51 |
4930512B01Rik |
RIKEN cDNA 4930512B01 gene |
6855 |
0.13 |
| chr14_25536740_25537143 | 0.51 |
Mir3075 |
microRNA 3075 |
2502 |
0.26 |
| chr16_8687825_8687976 | 0.51 |
Gm5767 |
predicted gene 5767 |
5070 |
0.14 |
| chr14_69282071_69282727 | 0.51 |
Gm20236 |
predicted gene, 20236 |
259 |
0.8 |
| chr14_69500320_69500976 | 0.50 |
Gm37094 |
predicted gene, 37094 |
258 |
0.81 |
| chr5_74064229_74066220 | 0.50 |
Usp46 |
ubiquitin specific peptidase 46 |
524 |
0.65 |
| chr15_73182349_73182927 | 0.49 |
Ago2 |
argonaute RISC catalytic subunit 2 |
2297 |
0.3 |
| chr12_118650955_118651106 | 0.49 |
Gm9267 |
predicted gene 9267 |
48353 |
0.15 |
| chr11_4487067_4487818 | 0.49 |
Mtmr3 |
myotubularin related protein 3 |
924 |
0.59 |
| chr3_95517184_95517335 | 0.48 |
Ctss |
cathepsin S |
9527 |
0.1 |
| chr14_118229781_118230155 | 0.48 |
Gm4675 |
predicted gene 4675 |
6264 |
0.13 |
| chr10_19483385_19484135 | 0.48 |
Gm33104 |
predicted gene, 33104 |
2955 |
0.29 |
| chr11_69399334_69402458 | 0.48 |
Kdm6bos |
KDM1 lysine (K)-specific demethylase 6B, opposite strand |
617 |
0.47 |
| chr6_89355649_89355800 | 0.47 |
Plxna1 |
plexin A1 |
1993 |
0.29 |
| chr10_78427526_78427677 | 0.47 |
4921516A02Rik |
RIKEN cDNA 4921516A02 gene |
670 |
0.41 |
| chr10_81153659_81153953 | 0.46 |
Pias4 |
protein inhibitor of activated STAT 4 |
2318 |
0.11 |
| chr17_15501788_15502060 | 0.46 |
Tbp |
TATA box binding protein |
1065 |
0.34 |
| chr3_154092419_154093223 | 0.46 |
Gm3076 |
predicted gene 3076 |
39519 |
0.13 |
| chr5_137290070_137291279 | 0.46 |
Ache |
acetylcholinesterase |
1427 |
0.17 |
| chr16_8670580_8670942 | 0.45 |
Carhsp1 |
calcium regulated heat stable protein 1 |
1394 |
0.29 |
| chr13_75715151_75715379 | 0.45 |
Ell2 |
elongation factor RNA polymerase II 2 |
7554 |
0.13 |
| chr13_23417840_23417991 | 0.45 |
Abt1 |
activator of basal transcription 1 |
5951 |
0.09 |
| chr18_54422355_54422833 | 0.44 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
299 |
0.93 |
| chr13_100676259_100676561 | 0.44 |
Ccdc125 |
coiled-coil domain containing 125 |
6637 |
0.12 |
| chr14_115046048_115046375 | 0.44 |
Mir92-1 |
microRNA 92-1 |
1784 |
0.14 |
| chr5_139391800_139391951 | 0.43 |
Gpr146 |
G protein-coupled receptor 146 |
2090 |
0.2 |
| chr11_94264019_94264170 | 0.43 |
Gm21885 |
predicted gene, 21885 |
17779 |
0.14 |
| chr15_103250315_103251530 | 0.43 |
Nfe2 |
nuclear factor, erythroid derived 2 |
543 |
0.62 |
| chr7_62414907_62415687 | 0.42 |
Mkrn3 |
makorin, ring finger protein, 3 |
4842 |
0.18 |
| chr1_90947541_90947692 | 0.42 |
Prlh |
prolactin releasing hormone |
5492 |
0.15 |
| chr10_127323007_127323393 | 0.42 |
Arhgap9 |
Rho GTPase activating protein 9 |
507 |
0.55 |
| chr11_4488078_4488309 | 0.42 |
Mtmr3 |
myotubularin related protein 3 |
173 |
0.95 |
| chr2_116056837_116059755 | 0.41 |
Meis2 |
Meis homeobox 2 |
546 |
0.75 |
| chr7_16103274_16103425 | 0.41 |
Napa |
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
4267 |
0.14 |
| chr11_116647941_116648526 | 0.41 |
Prcd |
photoreceptor disc component |
5301 |
0.09 |
| chr1_9780296_9780452 | 0.41 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
3404 |
0.17 |
| chr16_91222072_91224159 | 0.41 |
Gm49614 |
predicted gene, 49614 |
189 |
0.9 |
| chr4_108060098_108060443 | 0.40 |
Scp2 |
sterol carrier protein 2, liver |
11093 |
0.13 |
| chrX_42155222_42155373 | 0.40 |
Stag2 |
stromal antigen 2 |
3117 |
0.24 |
| chr10_57499477_57499901 | 0.40 |
Hsf2 |
heat shock factor 2 |
685 |
0.68 |
| chr5_137382579_137382730 | 0.40 |
Zan |
zonadhesin |
19084 |
0.09 |
| chr6_90333325_90334527 | 0.40 |
Uroc1 |
urocanase domain containing 1 |
637 |
0.57 |
| chr17_34950362_34951201 | 0.40 |
Snord52 |
small nucleolar RNA, C/D box 52 |
236 |
0.58 |
| chr4_154966258_154966899 | 0.39 |
Pank4 |
pantothenate kinase 4 |
2445 |
0.18 |
| chr16_8612946_8613812 | 0.39 |
Abat |
4-aminobutyrate aminotransferase |
501 |
0.67 |
| chr3_89209863_89210054 | 0.39 |
Mtx1 |
metaxin 1 |
136 |
0.74 |
| chr11_48803049_48803584 | 0.39 |
Rack1 |
receptor for activated C kinase 1 |
164 |
0.48 |
| chr6_88501831_88502191 | 0.39 |
Gm44170 |
predicted gene, 44170 |
1594 |
0.29 |
| chr8_71377836_71378740 | 0.39 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
2498 |
0.14 |
| chr12_103735553_103735704 | 0.39 |
Serpina1b |
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
2530 |
0.16 |
| chr6_41702817_41703169 | 0.39 |
Kel |
Kell blood group |
1346 |
0.32 |
| chr18_14459496_14459647 | 0.39 |
Gm50098 |
predicted gene, 50098 |
32434 |
0.17 |
| chr1_67125661_67125812 | 0.39 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
2710 |
0.34 |
| chr7_127661632_127661895 | 0.38 |
Gm44760 |
predicted gene 44760 |
3529 |
0.11 |
| chr13_73483496_73483773 | 0.38 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
11883 |
0.21 |
| chr2_152785455_152786223 | 0.38 |
Gm23802 |
predicted gene, 23802 |
12950 |
0.11 |
| chr7_19691393_19692666 | 0.38 |
Apoc1 |
apolipoprotein C-I |
462 |
0.59 |
| chr1_136960004_136960405 | 0.38 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
176 |
0.96 |
| chr19_14889594_14889745 | 0.38 |
Gm5513 |
predicted pseudogene 5513 |
71097 |
0.13 |
| chr11_96202439_96204563 | 0.38 |
Hoxb13 |
homeobox B13 |
9185 |
0.1 |
| chrX_150554562_150554911 | 0.37 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
699 |
0.62 |
| chrX_96241467_96241787 | 0.37 |
Mir223 |
microRNA 223 |
1190 |
0.35 |
| chr15_12203564_12203715 | 0.37 |
Mtmr12 |
myotubularin related protein 12 |
1389 |
0.28 |
| chr5_35609246_35609861 | 0.37 |
Acox3 |
acyl-Coenzyme A oxidase 3, pristanoyl |
546 |
0.71 |
| chr9_79823449_79823600 | 0.36 |
Filip1 |
filamin A interacting protein 1 |
7474 |
0.15 |
| chr1_168381191_168381418 | 0.36 |
Gm38381 |
predicted gene, 38381 |
30496 |
0.18 |
| chr13_59668566_59668950 | 0.36 |
Golm1 |
golgi membrane protein 1 |
7014 |
0.1 |
| chr14_98164357_98165375 | 0.36 |
Dach1 |
dachshund family transcription factor 1 |
4677 |
0.28 |
| chr9_70502649_70503336 | 0.36 |
Rnf111 |
ring finger 111 |
362 |
0.81 |
| chr4_154960570_154962200 | 0.36 |
Gm20421 |
predicted gene 20421 |
293 |
0.57 |
| chr12_29529828_29531185 | 0.36 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr1_121177384_121178460 | 0.36 |
Gm29359 |
predicted gene 29359 |
46354 |
0.15 |
| chr11_58868459_58868610 | 0.35 |
2810021J22Rik |
RIKEN cDNA 2810021J22 gene |
1267 |
0.21 |
| chr8_70656342_70656817 | 0.35 |
Pgpep1 |
pyroglutamyl-peptidase I |
859 |
0.37 |
| chr2_84735706_84738103 | 0.35 |
Ypel4 |
yippee like 4 |
2677 |
0.11 |
| chr14_61650328_61650516 | 0.35 |
Gm27017 |
predicted gene, 27017 |
1472 |
0.19 |
| chr16_17949655_17949806 | 0.35 |
Slc25a1 |
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
21511 |
0.09 |
| chr8_61515132_61515409 | 0.35 |
Palld |
palladin, cytoskeletal associated protein |
630 |
0.77 |
| chr15_96292843_96292994 | 0.34 |
Arid2 |
AT rich interactive domain 2 (ARID, RFX-like) |
5345 |
0.21 |
| chr11_3488055_3488462 | 0.34 |
Pla2g3 |
phospholipase A2, group III |
26 |
0.95 |
| chr1_71100990_71101619 | 0.34 |
Bard1 |
BRCA1 associated RING domain 1 |
1842 |
0.43 |
| chr15_76179745_76180476 | 0.34 |
Plec |
plectin |
7933 |
0.09 |
| chr11_60538080_60539333 | 0.34 |
Alkbh5 |
alkB homolog 5, RNA demethylase |
728 |
0.48 |
| chr8_47531539_47531690 | 0.34 |
Trappc11 |
trafficking protein particle complex 11 |
1853 |
0.25 |
| chr4_130108875_130109149 | 0.34 |
Pef1 |
penta-EF hand domain containing 1 |
1456 |
0.34 |
| chr5_64588253_64588408 | 0.34 |
Gm3716 |
predicted gene 3716 |
14830 |
0.11 |
| chr9_114562215_114562607 | 0.34 |
Trim71 |
tripartite motif-containing 71 |
1958 |
0.3 |
| chr2_33867715_33867866 | 0.34 |
Mvb12b |
multivesicular body subunit 12B |
19055 |
0.2 |
| chr3_108573576_108574011 | 0.33 |
Taf13 |
TATA-box binding protein associated factor 13 |
2082 |
0.15 |
| chr13_97778790_97779019 | 0.33 |
Gm47577 |
predicted gene, 47577 |
11660 |
0.15 |
| chr9_110653127_110654518 | 0.33 |
Nbeal2 |
neurobeachin-like 2 |
320 |
0.77 |
| chr13_83741336_83742027 | 0.33 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2818 |
0.16 |
| chr10_80834399_80836114 | 0.33 |
Mir1982 |
microRNA 1982 |
6459 |
0.07 |
| chr10_43580494_43581222 | 0.33 |
Cd24a |
CD24a antigen |
923 |
0.48 |
| chr11_55449682_55449833 | 0.33 |
Atox1 |
antioxidant 1 copper chaperone |
11440 |
0.11 |
| chr1_75235806_75235957 | 0.33 |
Dnajb2 |
DnaJ heat shock protein family (Hsp40) member B2 |
525 |
0.5 |
| chr18_54436033_54436373 | 0.32 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
13908 |
0.23 |
| chr5_34369874_34370083 | 0.32 |
Fam193a |
family with sequence homology 193, member A |
45 |
0.97 |
| chr17_26974792_26975099 | 0.32 |
Zbtb9 |
zinc finger and BTB domain containing 9 |
1728 |
0.14 |
| chr1_130732649_130733832 | 0.32 |
AA986860 |
expressed sequence AA986860 |
1130 |
0.29 |
| chr16_8687033_8687675 | 0.32 |
Gm5767 |
predicted gene 5767 |
4524 |
0.14 |
| chr10_81349455_81350420 | 0.32 |
Hmg20b |
high mobility group 20B |
254 |
0.74 |
| chr13_59763390_59763779 | 0.32 |
Isca1 |
iron-sulfur cluster assembly 1 |
17 |
0.94 |
| chr1_23282787_23283466 | 0.31 |
Gm27028 |
predicted gene, 27028 |
8411 |
0.12 |
| chr19_53332272_53332423 | 0.31 |
Mxi1 |
MAX interactor 1, dimerization protein |
1939 |
0.25 |
| chr2_105908509_105908660 | 0.31 |
Immp1l |
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
3946 |
0.21 |
| chr9_59660933_59662180 | 0.31 |
Pkm |
pyruvate kinase, muscle |
3246 |
0.17 |
| chr4_100995628_100995943 | 0.31 |
Cachd1 |
cache domain containing 1 |
1075 |
0.53 |
| chrX_153501207_153502250 | 0.31 |
Ubqln2 |
ubiquilin 2 |
3501 |
0.22 |
| chr5_112001842_112002220 | 0.31 |
Gm42488 |
predicted gene 42488 |
57796 |
0.13 |
| chr11_95013727_95013935 | 0.30 |
Samd14 |
sterile alpha motif domain containing 14 |
3550 |
0.13 |
| chr5_104043873_104044030 | 0.30 |
Nudt9 |
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
2355 |
0.16 |
| chr1_177150233_177151142 | 0.30 |
Gm38146 |
predicted gene, 38146 |
15234 |
0.16 |
| chr5_142589584_142589802 | 0.30 |
Mmd2 |
monocyte to macrophage differentiation-associated 2 |
6825 |
0.15 |
| chr9_107723003_107723309 | 0.30 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
12681 |
0.09 |
| chr1_139375797_139376462 | 0.30 |
Crb1 |
crumbs family member 1, photoreceptor morphogenesis associated |
912 |
0.52 |
| chr3_84219260_84219630 | 0.30 |
Trim2 |
tripartite motif-containing 2 |
1400 |
0.49 |
| chr3_27146047_27146198 | 0.30 |
Ect2 |
ect2 oncogene |
1079 |
0.48 |
| chr2_147180654_147181518 | 0.30 |
Nkx2-2os |
NK2 homeobox 2, opposite strand |
2997 |
0.2 |
| chr3_105054815_105055585 | 0.29 |
Cttnbp2nl |
CTTNBP2 N-terminal like |
2054 |
0.3 |
| chr11_57791079_57791485 | 0.29 |
2010001A14Rik |
RIKEN cDNA 2010001A14 gene |
10116 |
0.14 |
| chr16_30129996_30130199 | 0.29 |
Gm6029 |
predicted gene 6029 |
9362 |
0.15 |
| chr10_50891809_50893729 | 0.29 |
Sim1 |
single-minded family bHLH transcription factor 1 |
1985 |
0.43 |
| chrX_12067726_12070081 | 0.29 |
Bcor |
BCL6 interacting corepressor |
11650 |
0.26 |
| chr7_100492685_100494805 | 0.29 |
Ucp2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
50 |
0.95 |
| chr4_132274460_132275262 | 0.29 |
Taf12 |
TATA-box binding protein associated factor 12 |
445 |
0.57 |
| chr4_140704067_140704218 | 0.29 |
Rcc2 |
regulator of chromosome condensation 2 |
2669 |
0.17 |
| chr8_26384770_26384921 | 0.29 |
Gm31898 |
predicted gene, 31898 |
50868 |
0.1 |
| chr16_96441846_96442164 | 0.29 |
Itgb2l |
integrin beta 2-like |
1607 |
0.32 |
| chr11_102023682_102023894 | 0.29 |
Mpp3 |
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
2069 |
0.17 |
| chr1_161734865_161735433 | 0.28 |
Gm15429 |
predicted pseudogene 15429 |
7660 |
0.15 |
| chr8_92364963_92367136 | 0.28 |
Irx5 |
Iroquois homeobox 5 |
8300 |
0.18 |
| chr14_34373709_34374792 | 0.28 |
Sncg |
synuclein, gamma |
539 |
0.58 |
| chr15_76666348_76670076 | 0.28 |
Foxh1 |
forkhead box H1 |
1590 |
0.15 |
| chr5_117122436_117122623 | 0.28 |
Taok3 |
TAO kinase 3 |
2400 |
0.21 |
| chr6_31124166_31124436 | 0.28 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
13381 |
0.11 |
| chr1_13574325_13574563 | 0.28 |
Tram1 |
translocating chain-associating membrane protein 1 |
5354 |
0.24 |
| chr6_122609501_122610041 | 0.28 |
Gdf3 |
growth differentiation factor 3 |
53 |
0.95 |
| chr11_106312219_106312370 | 0.28 |
Cd79b |
CD79B antigen |
2235 |
0.17 |
| chr11_100627464_100627615 | 0.28 |
Nkiras2 |
NFKB inhibitor interacting Ras-like protein 2 |
4528 |
0.11 |
| chr12_91747422_91747591 | 0.27 |
Ston2 |
stonin 2 |
1422 |
0.4 |
| chr11_45855533_45856802 | 0.27 |
Clint1 |
clathrin interactor 1 |
4203 |
0.18 |
| chr19_55285309_55285619 | 0.27 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
1130 |
0.48 |
| chr14_30942231_30942467 | 0.27 |
Itih1 |
inter-alpha trypsin inhibitor, heavy chain 1 |
928 |
0.4 |
| chr15_98843609_98844032 | 0.27 |
Kmt2d |
lysine (K)-specific methyltransferase 2D |
5417 |
0.09 |
| chr3_68695313_68695505 | 0.27 |
Il12a |
interleukin 12a |
1388 |
0.42 |
| chr4_53633406_53634189 | 0.27 |
Fsd1l |
fibronectin type III and SPRY domain containing 1-like |
2081 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 0.9 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.2 | 0.8 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.2 | 1.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 0.5 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.6 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 1.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:2000981 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.2 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.2 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 1.1 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.7 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0071864 | positive regulation of cell proliferation in bone marrow(GO:0071864) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:1904220 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.0 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.0 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 0.8 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.6 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.8 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0018593 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0008932 | lytic endotransglycosylase activity(GO:0008932) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |