Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
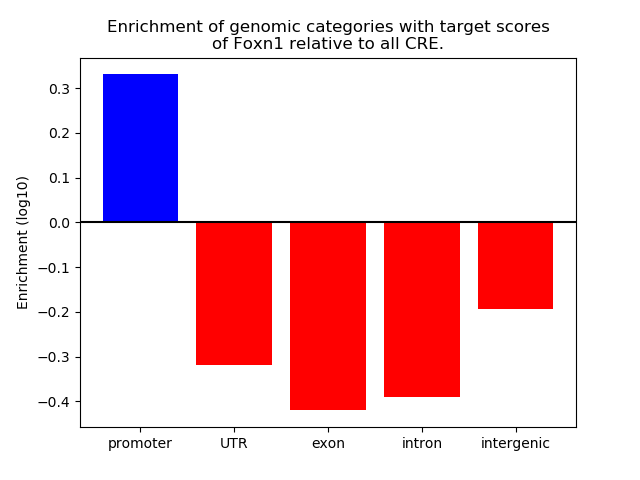
Results for Foxn1
Z-value: 0.43
Transcription factors associated with Foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxn1
|
ENSMUSG00000002057.4 | forkhead box N1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_78384887_78385288 | Foxn1 | 1471 | 0.236917 | -0.22 | 9.8e-02 | Click! |
| chr11_78386075_78386548 | Foxn1 | 247 | 0.847600 | -0.20 | 1.2e-01 | Click! |
| chr11_78383526_78383677 | Foxn1 | 2957 | 0.133104 | -0.15 | 2.5e-01 | Click! |
| chr11_78378001_78378152 | Foxn1 | 8482 | 0.094759 | -0.08 | 5.3e-01 | Click! |
| chr11_78385731_78386019 | Foxn1 | 683 | 0.508496 | -0.06 | 6.2e-01 | Click! |
Activity of the Foxn1 motif across conditions
Conditions sorted by the z-value of the Foxn1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
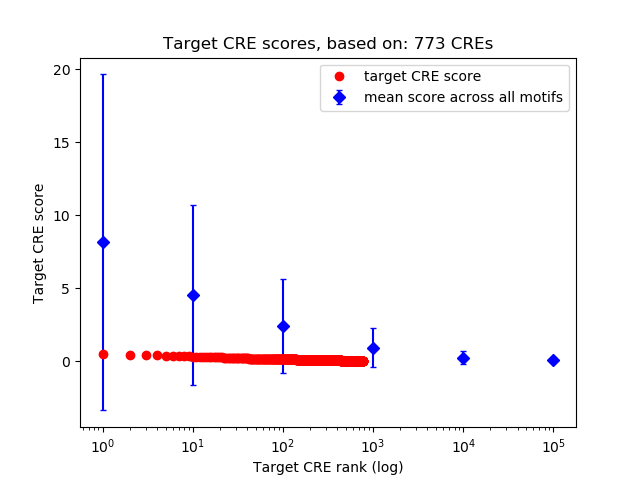
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_83721518_83722206 | 0.52 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
481 |
0.73 |
| chrX_23285148_23285871 | 0.43 |
Klhl13 |
kelch-like 13 |
50 |
0.99 |
| chr10_34301557_34301950 | 0.42 |
Tspyl4 |
TSPY-like 4 |
2497 |
0.16 |
| chr9_96758931_96759244 | 0.41 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
6256 |
0.16 |
| chr5_22746211_22746970 | 0.38 |
A930003O13Rik |
RIKEN cDNA A930003O13 gene |
297 |
0.65 |
| chr9_113708285_113708476 | 0.38 |
Pdcd6ip |
programmed cell death 6 interacting protein |
121 |
0.97 |
| chr2_20966052_20966447 | 0.35 |
Arhgap21 |
Rho GTPase activating protein 21 |
1471 |
0.39 |
| chr13_13785297_13785737 | 0.35 |
Gng4 |
guanine nucleotide binding protein (G protein), gamma 4 |
1188 |
0.47 |
| chr8_123333278_123333633 | 0.33 |
Spire2 |
spire type actin nucleation factor 2 |
742 |
0.47 |
| chr2_126675916_126676238 | 0.32 |
Gm27003 |
predicted gene, 27003 |
161 |
0.62 |
| chr14_70577418_70577686 | 0.32 |
Nudt18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
85 |
0.95 |
| chr7_36705087_36705327 | 0.31 |
Gm37452 |
predicted gene, 37452 |
4967 |
0.15 |
| chr10_40257841_40258099 | 0.31 |
Gtf3c6 |
general transcription factor IIIC, polypeptide 6, alpha |
138 |
0.81 |
| chr19_27217816_27218252 | 0.31 |
Vldlr |
very low density lipoprotein receptor |
576 |
0.77 |
| chr6_105676979_105678296 | 0.29 |
Cntn4 |
contactin 4 |
23 |
0.52 |
| chr9_62040106_62040360 | 0.29 |
Paqr5 |
progestin and adipoQ receptor family member V |
13377 |
0.2 |
| chr6_121183940_121184503 | 0.29 |
Pex26 |
peroxisomal biogenesis factor 26 |
323 |
0.84 |
| chr1_188003839_188004226 | 0.28 |
9330162B11Rik |
RIKEN cDNA 9330162B11 gene |
4958 |
0.26 |
| chr8_104629629_104629943 | 0.27 |
Gm33023 |
predicted gene, 33023 |
1046 |
0.27 |
| chr18_11657869_11658260 | 0.27 |
Rbbp8 |
retinoblastoma binding protein 8, endonuclease |
222 |
0.57 |
| chr8_87539918_87540560 | 0.27 |
4933402J07Rik |
RIKEN cDNA 4933402J07 gene |
23614 |
0.14 |
| chr14_55055435_55055869 | 0.26 |
Gm20687 |
predicted gene 20687 |
159 |
0.87 |
| chr17_34295833_34296830 | 0.26 |
H2-Aa |
histocompatibility 2, class II antigen A, alpha |
8508 |
0.08 |
| chr12_5373600_5373751 | 0.26 |
Klhl29 |
kelch-like 29 |
2007 |
0.33 |
| chr17_55447566_55447717 | 0.25 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
1397 |
0.51 |
| chr17_28176724_28177079 | 0.23 |
Zfp523 |
zinc finger protein 523 |
228 |
0.79 |
| chr9_35558084_35558540 | 0.23 |
Ddx25 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25 |
216 |
0.55 |
| chr16_38741987_38742274 | 0.23 |
B4galt4 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 4 |
134 |
0.95 |
| chr19_53661907_53662408 | 0.23 |
Rbm20 |
RNA binding motif protein 20 |
15149 |
0.16 |
| chr5_145191000_145191535 | 0.22 |
Atp5j2 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
244 |
0.85 |
| chr1_180387684_180387967 | 0.22 |
Gm36933 |
predicted gene, 36933 |
11381 |
0.14 |
| chr14_80901423_80902185 | 0.22 |
Gm49044 |
predicted gene, 49044 |
28933 |
0.21 |
| chr2_51753080_51753275 | 0.22 |
Gm13490 |
predicted gene 13490 |
21168 |
0.21 |
| chr5_80706733_80706963 | 0.22 |
Gm22974 |
predicted gene, 22974 |
186806 |
0.03 |
| chr9_78447914_78448215 | 0.21 |
Mto1 |
mitochondrial tRNA translation optimization 1 |
144 |
0.85 |
| chr5_120435825_120436608 | 0.21 |
Gm27199 |
predicted gene 27199 |
4449 |
0.14 |
| chrX_135210129_135210918 | 0.20 |
Tceal6 |
transcription elongation factor A (SII)-like 6 |
164 |
0.93 |
| chr9_75313853_75314949 | 0.20 |
Gnb5 |
guanine nucleotide binding protein (G protein), beta 5 |
260 |
0.66 |
| chr6_85587353_85587505 | 0.20 |
Alms1 |
ALMS1, centrosome and basal body associated |
102 |
0.96 |
| chr2_127008729_127009233 | 0.19 |
Ap4e1 |
adaptor-related protein complex AP-4, epsilon 1 |
227 |
0.93 |
| chr1_171360132_171360778 | 0.19 |
Klhdc9 |
kelch domain containing 9 |
343 |
0.67 |
| chr14_18271212_18271647 | 0.19 |
Rpl15 |
ribosomal protein L15 |
38 |
0.66 |
| chr7_127470523_127470867 | 0.18 |
Prr14 |
proline rich 14 |
324 |
0.69 |
| chr11_70239069_70239790 | 0.18 |
Rnasek |
ribonuclease, RNase K |
351 |
0.42 |
| chr13_64312809_64313057 | 0.18 |
Prxl2c |
peroxiredoxin like 2C |
223 |
0.87 |
| chr14_111677526_111678136 | 0.18 |
Slitrk5 |
SLIT and NTRK-like family, member 5 |
1982 |
0.36 |
| chr16_16896578_16897698 | 0.18 |
Ppm1f |
protein phosphatase 1F (PP2C domain containing) |
564 |
0.59 |
| chr7_4137072_4137259 | 0.18 |
Leng8 |
leukocyte receptor cluster (LRC) member 8 |
81 |
0.7 |
| chr2_128125371_128125665 | 0.18 |
Bcl2l11 |
BCL2-like 11 (apoptosis facilitator) |
520 |
0.81 |
| chr13_77046468_77046715 | 0.17 |
Slf1 |
SMC5-SMC6 complex localization factor 1 |
806 |
0.74 |
| chr18_69563283_69563781 | 0.17 |
Tcf4 |
transcription factor 4 |
197 |
0.96 |
| chr16_50689551_50689950 | 0.17 |
Gm9575 |
predicted gene 9575 |
23404 |
0.17 |
| chr17_6476493_6476766 | 0.17 |
Tmem181b-ps |
transmembrane protein 181B, pseudogene |
9607 |
0.17 |
| chr7_12949824_12950412 | 0.17 |
1810019N24Rik |
RIKEN cDNA 1810019N24 gene |
52 |
0.93 |
| chr10_33083500_33083959 | 0.17 |
Trdn |
triadin |
168 |
0.97 |
| chr11_121014589_121015240 | 0.17 |
Gm11774 |
predicted gene 11774 |
11130 |
0.09 |
| chr19_5913025_5913204 | 0.17 |
Dpf2 |
D4, zinc and double PHD fingers family 2 |
104 |
0.51 |
| chr3_122274436_122274925 | 0.17 |
Dnttip2 |
deoxynucleotidyltransferase, terminal, interacting protein 2 |
262 |
0.83 |
| chr17_80061806_80062276 | 0.17 |
Hnrnpll |
heterogeneous nuclear ribonucleoprotein L-like |
94 |
0.96 |
| chr7_74013813_74014080 | 0.17 |
St8sia2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
256 |
0.92 |
| chr11_94993223_94993531 | 0.17 |
Ppp1r9b |
protein phosphatase 1, regulatory subunit 9B |
497 |
0.67 |
| chr7_57386487_57387529 | 0.17 |
Gabrg3 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 |
137 |
0.68 |
| chr11_53707522_53707690 | 0.17 |
Rad50 |
RAD50 double strand break repair protein |
287 |
0.84 |
| chr2_30285852_30286279 | 0.17 |
Dolk |
dolichol kinase |
269 |
0.52 |
| chr6_113482507_113483177 | 0.17 |
Creld1 |
cysteine-rich with EGF-like domains 1 |
455 |
0.61 |
| chr12_112644112_112644465 | 0.16 |
Siva1 |
SIVA1, apoptosis-inducing factor |
391 |
0.74 |
| chrX_74085086_74085553 | 0.16 |
Mecp2 |
methyl CpG binding protein 2 |
278 |
0.86 |
| chr9_13297922_13298581 | 0.16 |
Maml2 |
mastermind like transcriptional coactivator 2 |
294 |
0.88 |
| chr9_42264436_42264649 | 0.16 |
Sc5d |
sterol-C5-desaturase |
286 |
0.9 |
| chr3_52180562_52180887 | 0.16 |
Gm30074 |
predicted gene, 30074 |
752 |
0.55 |
| chr8_17534365_17535810 | 0.16 |
Csmd1 |
CUB and Sushi multiple domains 1 |
194 |
0.97 |
| chr1_57429727_57430347 | 0.16 |
Gm22371 |
predicted gene, 22371 |
7664 |
0.15 |
| chr3_62603065_62604257 | 0.16 |
Gpr149 |
G protein-coupled receptor 149 |
887 |
0.72 |
| chr7_125445497_125445648 | 0.16 |
Kdm8 |
lysine (K)-specific demethylase 8 |
888 |
0.57 |
| chr18_65490174_65490449 | 0.16 |
Gm50246 |
predicted gene, 50246 |
11095 |
0.13 |
| chr2_180953875_180955091 | 0.16 |
Gm14341 |
predicted gene 14341 |
19 |
0.62 |
| chr8_63951776_63952541 | 0.16 |
Sgo2b |
shugoshin 2B |
12 |
0.98 |
| chr11_43834010_43835381 | 0.16 |
Adra1b |
adrenergic receptor, alpha 1b |
1637 |
0.47 |
| chr3_158561890_158562423 | 0.15 |
Lrrc7 |
leucine rich repeat containing 7 |
22 |
0.99 |
| chr1_93511429_93512004 | 0.15 |
Farp2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
363 |
0.82 |
| chr8_113635587_113636301 | 0.15 |
Mon1b |
MON1 homolog B, secretory traffciking associated |
109 |
0.96 |
| chr6_38552191_38552670 | 0.15 |
Luc7l2 |
LUC7-like 2 (S. cerevisiae) |
587 |
0.68 |
| chr2_181864457_181865175 | 0.15 |
Polr3k |
polymerase (RNA) III (DNA directed) polypeptide K |
456 |
0.79 |
| chr6_112929880_112930575 | 0.15 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
16527 |
0.14 |
| chr17_46856362_46856796 | 0.15 |
Bicral |
BRD4 interacting chromatin remodeling complex associated protein like |
1903 |
0.28 |
| chr8_70896352_70896974 | 0.15 |
Rpl18a |
ribosomal protein L18A |
620 |
0.33 |
| chr16_32658548_32658939 | 0.15 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
182 |
0.93 |
| chr4_13754294_13755025 | 0.15 |
Runx1t1 |
RUNX1 translocation partner 1 |
3362 |
0.37 |
| chr9_47533054_47533827 | 0.15 |
Cadm1 |
cell adhesion molecule 1 |
3067 |
0.25 |
| chr15_44787203_44788294 | 0.15 |
A930017M01Rik |
RIKEN cDNA A930017M01 gene |
21 |
0.83 |
| chr9_27027276_27028031 | 0.15 |
Vps26b |
VPS26 retromer complex component B |
2377 |
0.17 |
| chr2_64853261_64853765 | 0.15 |
Gm13578 |
predicted gene 13578 |
15075 |
0.27 |
| chr15_27467790_27468148 | 0.15 |
Ank |
progressive ankylosis |
1292 |
0.39 |
| chr5_120711312_120711715 | 0.14 |
Dtx1 |
deltex 1, E3 ubiquitin ligase |
414 |
0.73 |
| chr14_26639283_26639494 | 0.14 |
Arf4 |
ADP-ribosylation factor 4 |
1041 |
0.32 |
| chr4_155891860_155892180 | 0.14 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
118 |
0.64 |
| chr1_55088037_55088229 | 0.14 |
Hspe1 |
heat shock protein 1 (chaperonin 10) |
1 |
0.59 |
| chr11_41999400_42000640 | 0.14 |
Gabrg2 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
336 |
0.92 |
| chrX_102020230_102020400 | 0.14 |
Gm27843 |
predicted gene, 27843 |
7163 |
0.16 |
| chr11_100738402_100738693 | 0.14 |
Rab5c |
RAB5C, member RAS oncogene family |
332 |
0.77 |
| chr15_72807706_72808811 | 0.14 |
Peg13 |
paternally expressed 13 |
2066 |
0.4 |
| chr17_25239740_25240111 | 0.14 |
Gnptg |
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
191 |
0.53 |
| chr19_6241712_6242757 | 0.14 |
Atg2a |
autophagy related 2A |
566 |
0.52 |
| chr7_36604988_36605635 | 0.14 |
Gm29129 |
predicted gene 29129 |
36568 |
0.17 |
| chr4_82509049_82510307 | 0.14 |
Gm11266 |
predicted gene 11266 |
1662 |
0.38 |
| chr4_43499187_43499622 | 0.14 |
Arhgef39 |
Rho guanine nucleotide exchange factor (GEF) 39 |
204 |
0.84 |
| chr8_87974200_87975457 | 0.14 |
Zfp423 |
zinc finger protein 423 |
15233 |
0.24 |
| chr11_101641133_101641480 | 0.14 |
Gm24950 |
predicted gene, 24950 |
442 |
0.64 |
| chr11_46057942_46058389 | 0.14 |
Adam19 |
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
2107 |
0.23 |
| chr12_9029981_9031052 | 0.14 |
Ttc32 |
tetratricopeptide repeat domain 32 |
488 |
0.82 |
| chr4_13749333_13749927 | 0.14 |
Runx1t1 |
RUNX1 translocation partner 1 |
1667 |
0.52 |
| chr12_113140557_113141515 | 0.14 |
Crip2 |
cysteine rich protein 2 |
464 |
0.68 |
| chr13_48261032_48261213 | 0.14 |
Id4 |
inhibitor of DNA binding 4 |
106 |
0.92 |
| chr11_84915695_84916573 | 0.14 |
Znhit3 |
zinc finger, HIT type 3 |
171 |
0.93 |
| chr11_70240387_70240647 | 0.14 |
Rnasek |
ribonuclease, RNase K |
675 |
0.3 |
| chr14_51089768_51090539 | 0.13 |
Rnase4 |
ribonuclease, RNase A family 4 |
924 |
0.27 |
| chr6_128803150_128803507 | 0.13 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
14113 |
0.09 |
| chr8_84102100_84102251 | 0.13 |
Dcaf15 |
DDB1 and CUL4 associated factor 15 |
2587 |
0.11 |
| chr7_47050631_47051464 | 0.13 |
Tmem86a |
transmembrane protein 86A |
446 |
0.55 |
| chr6_7693227_7693490 | 0.13 |
Asns |
asparagine synthetase |
104 |
0.97 |
| chr13_77975252_77975403 | 0.13 |
Pou5f2 |
POU domain class 5, transcription factor 2 |
49575 |
0.15 |
| chr3_131564120_131564526 | 0.13 |
Papss1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
445 |
0.87 |
| chr10_81192796_81193377 | 0.13 |
Atcayos |
ataxia, cerebellar, Cayman type, opposite strand |
1523 |
0.14 |
| chr3_51415620_51415911 | 0.13 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
251 |
0.84 |
| chr4_122958691_122959270 | 0.13 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
1010 |
0.42 |
| chr3_95928548_95928975 | 0.13 |
Anp32e |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
485 |
0.63 |
| chr1_77501424_77502014 | 0.13 |
Mir6352 |
microRNA 6352 |
4951 |
0.21 |
| chr19_45812760_45812972 | 0.13 |
Kcnip2 |
Kv channel-interacting protein 2 |
575 |
0.7 |
| chr5_108629016_108629687 | 0.13 |
Gak |
cyclin G associated kinase |
72 |
0.84 |
| chr16_94996546_94998023 | 0.13 |
Kcnj6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
32 |
0.99 |
| chr4_102254418_102255744 | 0.13 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
78 |
0.99 |
| chr15_83172415_83172866 | 0.12 |
Cyb5r3 |
cytochrome b5 reductase 3 |
48 |
0.78 |
| chr6_113442622_113443115 | 0.12 |
Jagn1 |
jagunal homolog 1 |
258 |
0.77 |
| chr17_56141491_56142567 | 0.12 |
Sema6b |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
1686 |
0.19 |
| chr5_111421306_111422790 | 0.12 |
Gm43119 |
predicted gene 43119 |
1541 |
0.35 |
| chr7_112954380_112955414 | 0.12 |
Rassf10 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 |
935 |
0.58 |
| chr1_65178523_65179140 | 0.12 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
350 |
0.84 |
| chr2_66408526_66409644 | 0.12 |
Gm13630 |
predicted gene 13630 |
409 |
0.72 |
| chr7_79148061_79149306 | 0.12 |
Mfge8 |
milk fat globule-EGF factor 8 protein |
306 |
0.89 |
| chr11_101646166_101646629 | 0.12 |
Gm23971 |
predicted gene, 23971 |
467 |
0.62 |
| chr16_32099114_32099696 | 0.12 |
Pigx |
phosphatidylinositol glycan anchor biosynthesis, class X |
288 |
0.56 |
| chr6_97938208_97938822 | 0.12 |
Mitf |
melanogenesis associated transcription factor |
2403 |
0.38 |
| chr6_122309029_122310024 | 0.12 |
M6pr |
mannose-6-phosphate receptor, cation dependent |
527 |
0.67 |
| chr11_98981811_98983058 | 0.12 |
Gjd3 |
gap junction protein, delta 3 |
582 |
0.6 |
| chr6_121184526_121184909 | 0.12 |
Pex26 |
peroxisomal biogenesis factor 26 |
819 |
0.52 |
| chr11_47988311_47989415 | 0.12 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
106 |
0.98 |
| chr16_11176493_11176867 | 0.12 |
Zc3h7a |
zinc finger CCCH type containing 7 A |
287 |
0.77 |
| chr4_156035263_156035900 | 0.12 |
Gm16008 |
predicted gene 16008 |
3938 |
0.08 |
| chr12_83987542_83988596 | 0.12 |
Acot2 |
acyl-CoA thioesterase 2 |
208 |
0.87 |
| chr18_3507019_3507669 | 0.12 |
Bambi |
BMP and activin membrane-bound inhibitor |
579 |
0.7 |
| chrX_12916494_12917181 | 0.12 |
Gm14524 |
predicted gene 14524 |
2245 |
0.27 |
| chr4_86734155_86734345 | 0.12 |
Dennd4c |
DENN/MADD domain containing 4C |
14305 |
0.2 |
| chr17_8368706_8369049 | 0.12 |
T2 |
brachyury 2 |
3519 |
0.15 |
| chr9_122571597_122571748 | 0.11 |
9530059O14Rik |
RIKEN cDNA 9530059O14 gene |
827 |
0.55 |
| chr11_69991633_69992222 | 0.11 |
Gabarap |
gamma-aminobutyric acid receptor associated protein |
120 |
0.87 |
| chr18_66291045_66291813 | 0.11 |
Ccbe1 |
collagen and calcium binding EGF domains 1 |
17 |
0.66 |
| chr13_99724294_99724631 | 0.11 |
Gm24471 |
predicted gene, 24471 |
31933 |
0.2 |
| chr4_120715702_120717192 | 0.11 |
Kcnq4 |
potassium voltage-gated channel, subfamily Q, member 4 |
30801 |
0.11 |
| chr11_60178073_60178451 | 0.11 |
Rai1 |
retinoic acid induced 1 |
2383 |
0.2 |
| chr8_86624092_86625039 | 0.11 |
Lonp2 |
lon peptidase 2, peroxisomal |
460 |
0.8 |
| chr2_164910537_164911246 | 0.11 |
Zfp335os |
zinc finger protein 335, opposite strand |
463 |
0.57 |
| chr15_81872520_81873197 | 0.11 |
Aco2 |
aconitase 2, mitochondrial |
168 |
0.85 |
| chr1_189921588_189922055 | 0.11 |
Smyd2 |
SET and MYND domain containing 2 |
432 |
0.82 |
| chr3_8923882_8924255 | 0.11 |
Mrps28 |
mitochondrial ribosomal protein S28 |
150 |
0.96 |
| chr3_70874375_70874526 | 0.11 |
Gm10780 |
predicted gene 10780 |
185358 |
0.03 |
| chr13_43158853_43159330 | 0.11 |
Tbc1d7 |
TBC1 domain family, member 7 |
217 |
0.94 |
| chr11_116652562_116653193 | 0.11 |
Prcd |
photoreceptor disc component |
657 |
0.45 |
| chr5_111429061_111430219 | 0.11 |
Gm43119 |
predicted gene 43119 |
6051 |
0.18 |
| chr11_116586983_116587414 | 0.11 |
Aanat |
arylalkylamine N-acetyltransferase |
766 |
0.46 |
| chr4_82489730_82490472 | 0.11 |
Nfib |
nuclear factor I/B |
9215 |
0.21 |
| chr7_133776817_133777117 | 0.11 |
Fank1 |
fibronectin type 3 and ankyrin repeat domains 1 |
25 |
0.67 |
| chr8_14384129_14384280 | 0.11 |
Dlgap2 |
DLG associated protein 2 |
2208 |
0.37 |
| chr2_71983646_71983797 | 0.11 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
2434 |
0.28 |
| chr7_110163999_110164879 | 0.11 |
1600010M07Rik |
RIKEN cDNA 1600010M07 gene |
13237 |
0.15 |
| chr4_137388448_137388745 | 0.11 |
2810405F17Rik |
RIKEN cDNA 2810405F17 gene |
214 |
0.89 |
| chr11_50210297_50211093 | 0.10 |
Sqstm1 |
sequestosome 1 |
70 |
0.67 |
| chr2_116063125_116063508 | 0.10 |
Meis2 |
Meis homeobox 2 |
786 |
0.59 |
| chr11_76763713_76763912 | 0.10 |
Gosr1 |
golgi SNAP receptor complex member 1 |
233 |
0.93 |
| chr2_152395546_152396207 | 0.10 |
Sox12 |
SRY (sex determining region Y)-box 12 |
2170 |
0.15 |
| chr1_93100315_93101980 | 0.10 |
Kif1a |
kinesin family member 1A |
675 |
0.62 |
| chr11_28365090_28365896 | 0.10 |
Gm12081 |
predicted gene 12081 |
5131 |
0.26 |
| chr1_139929330_139929725 | 0.10 |
Gm5837 |
predicted gene 5837 |
81 |
0.97 |
| chr2_153071959_153072358 | 0.10 |
Ccm2l |
cerebral cavernous malformation 2-like |
6156 |
0.15 |
| chr2_174463176_174464054 | 0.10 |
Atp5e |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
434 |
0.73 |
| chr1_95666998_95667591 | 0.10 |
St8sia4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
241 |
0.93 |
| chr10_5824069_5824916 | 0.10 |
Mtrf1l |
mitochondrial translational release factor 1-like |
582 |
0.79 |
| chr9_96733821_96734335 | 0.10 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
1308 |
0.4 |
| chr13_54621866_54622790 | 0.10 |
Faf2 |
Fas associated factor family member 2 |
490 |
0.69 |
| chr7_141228533_141229727 | 0.10 |
Phrf1 |
PHD and ring finger domains 1 |
341 |
0.71 |
| chr18_37496992_37497992 | 0.10 |
Pcdhb19 |
protocadherin beta 19 |
501 |
0.54 |
| chr1_190911845_190912265 | 0.10 |
Rps6kc1 |
ribosomal protein S6 kinase polypeptide 1 |
285 |
0.89 |
| chr19_3768976_3769359 | 0.10 |
Kmt5b |
lysine methyltransferase 5B |
620 |
0.58 |
| chr11_3451898_3452279 | 0.10 |
Rnf185 |
ring finger protein 185 |
246 |
0.5 |
| chr9_72985588_72986210 | 0.10 |
Ccpg1 |
cell cycle progression 1 |
280 |
0.62 |
| chr3_105053107_105053623 | 0.10 |
Cttnbp2nl |
CTTNBP2 N-terminal like |
219 |
0.93 |
| chr1_152399580_152399796 | 0.10 |
Colgalt2 |
collagen beta(1-O)galactosyltransferase 2 |
142 |
0.96 |
| chr19_49741229_49741380 | 0.10 |
Gm6975 |
predicted gene 6975 |
31180 |
0.19 |
| chr3_57735567_57735933 | 0.10 |
Rnf13 |
ring finger protein 13 |
312 |
0.85 |
| chr13_51848386_51848617 | 0.10 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
1757 |
0.42 |
| chr17_34204505_34204720 | 0.10 |
Tap2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
45 |
0.88 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.3 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.2 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |