Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
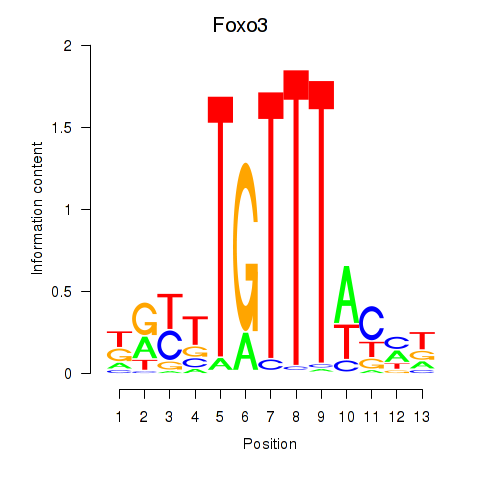
Results for Foxo3
Z-value: 0.55
Transcription factors associated with Foxo3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo3
|
ENSMUSG00000048756.5 | forkhead box O3 |
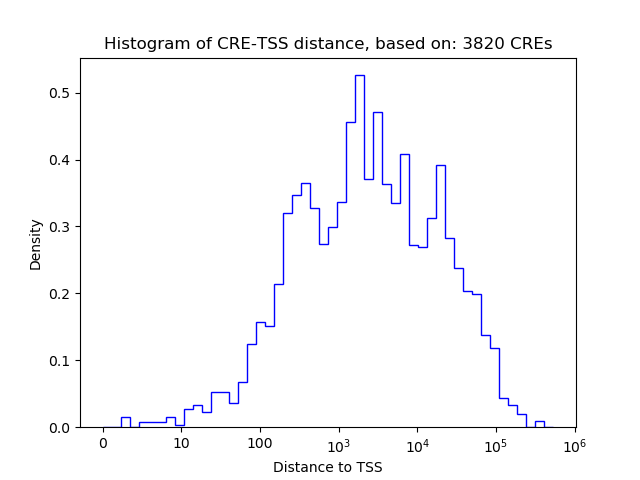
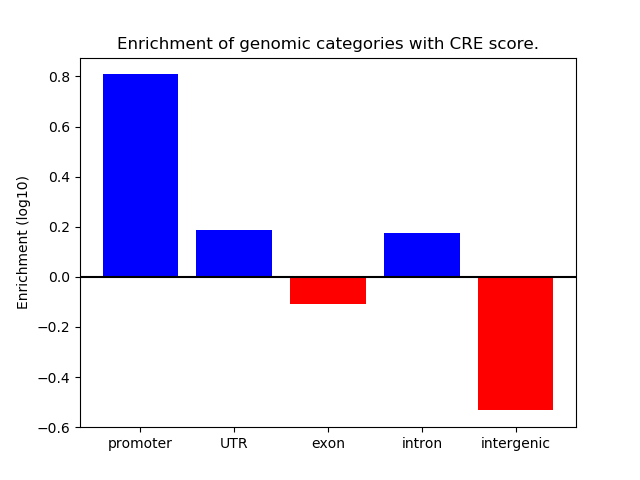
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_42272287_42272486 | Foxo3 | 4310 | 0.288598 | -0.39 | 2.1e-03 | Click! |
| chr10_42244059_42244384 | Foxo3 | 14145 | 0.250140 | -0.31 | 1.6e-02 | Click! |
| chr10_42251868_42252252 | Foxo3 | 6306 | 0.275620 | -0.31 | 1.6e-02 | Click! |
| chr10_42247495_42248433 | Foxo3 | 10402 | 0.259004 | -0.30 | 1.9e-02 | Click! |
| chr10_42262321_42262472 | Foxo3 | 4030 | 0.300609 | -0.30 | 1.9e-02 | Click! |
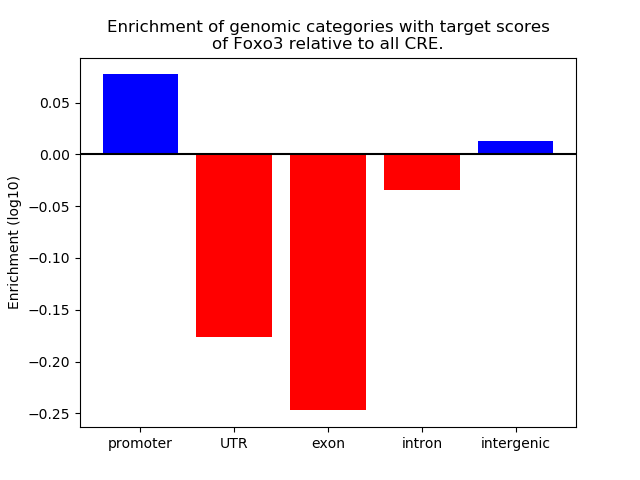
Activity of the Foxo3 motif across conditions
Conditions sorted by the z-value of the Foxo3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
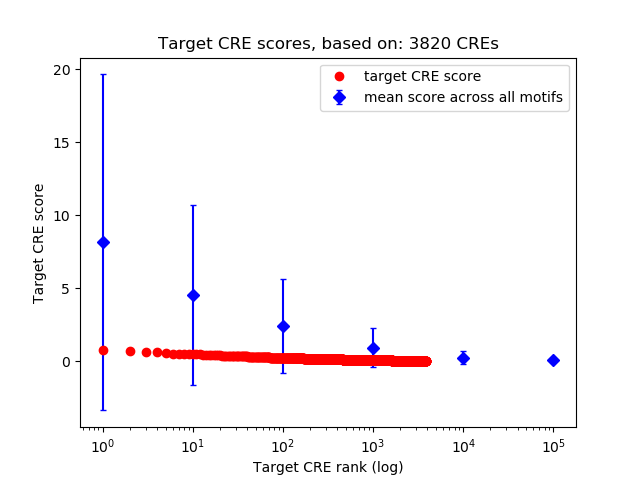
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_150261018_150262108 | 0.78 |
Fry |
FRY microtubule binding protein |
1796 |
0.34 |
| chr5_150260534_150260992 | 0.71 |
Fry |
FRY microtubule binding protein |
996 |
0.55 |
| chr8_25521556_25521707 | 0.60 |
Fgfr1 |
fibroblast growth factor receptor 1 |
1794 |
0.24 |
| chr18_69521278_69522871 | 0.60 |
Tcf4 |
transcription factor 4 |
73 |
0.98 |
| chr8_23669376_23670424 | 0.54 |
Zmat4 |
zinc finger, matrin type 4 |
226 |
0.95 |
| chr5_3432895_3433605 | 0.53 |
Cdk6 |
cyclin-dependent kinase 6 |
37699 |
0.12 |
| chr8_84765171_84766430 | 0.52 |
Nfix |
nuclear factor I/X |
7596 |
0.11 |
| chr6_55617797_55618517 | 0.51 |
Neurod6 |
neurogenic differentiation 6 |
63106 |
0.11 |
| chr6_8352545_8353207 | 0.49 |
Gm16055 |
predicted gene 16055 |
11260 |
0.18 |
| chr2_4300456_4301144 | 0.49 |
Frmd4a |
FERM domain containing 4A |
85 |
0.78 |
| chr5_57728215_57728366 | 0.49 |
Gm42635 |
predicted gene 42635 |
3897 |
0.15 |
| chr7_12421647_12422940 | 0.47 |
Zfp551 |
zinc fingr protein 551 |
146 |
0.92 |
| chr18_49543238_49543945 | 0.46 |
1700044K03Rik |
RIKEN cDNA 1700044K03 gene |
20302 |
0.23 |
| chr13_4662525_4663068 | 0.46 |
Gm40658 |
predicted gene, 40658 |
18889 |
0.18 |
| chr3_82141940_82142375 | 0.44 |
Gucy1a1 |
guanylate cyclase 1, soluble, alpha 1 |
2916 |
0.3 |
| chr6_136167149_136168437 | 0.42 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
4096 |
0.25 |
| chr7_144132849_144134003 | 0.42 |
Gm44999 |
predicted gene 44999 |
22341 |
0.2 |
| chr2_27539183_27539675 | 0.41 |
Gm25541 |
predicted gene, 25541 |
501 |
0.63 |
| chr7_30291688_30292446 | 0.41 |
Clip3 |
CAP-GLY domain containing linker protein 3 |
109 |
0.9 |
| chr11_109649280_109649701 | 0.41 |
Prkar1a |
protein kinase, cAMP dependent regulatory, type I, alpha |
85 |
0.97 |
| chr5_57721137_57722906 | 0.40 |
Pcdh7 |
protocadherin 7 |
108 |
0.94 |
| chr3_63296309_63296634 | 0.39 |
Mme |
membrane metallo endopeptidase |
376 |
0.91 |
| chr2_20339506_20340619 | 0.38 |
Etl4 |
enhancer trap locus 4 |
238 |
0.94 |
| chr1_153328595_153328920 | 0.38 |
Lamc1 |
laminin, gamma 1 |
4029 |
0.2 |
| chr5_107167246_107168359 | 0.38 |
Gm43423 |
predicted gene 43423 |
7231 |
0.17 |
| chr9_10903843_10905236 | 0.38 |
Cntn5 |
contactin 5 |
90 |
0.65 |
| chr11_93818459_93818772 | 0.36 |
Utp18 |
UTP18 small subunit processome component |
57528 |
0.12 |
| chr18_69125035_69125283 | 0.36 |
Gm23536 |
predicted gene, 23536 |
48961 |
0.13 |
| chr13_44946033_44946635 | 0.36 |
Dtnbp1 |
dystrobrevin binding protein 1 |
810 |
0.7 |
| chr1_23490694_23490976 | 0.36 |
Gm7784 |
predicted gene 7784 |
11296 |
0.24 |
| chr14_50896223_50897440 | 0.35 |
Klhl33 |
kelch-like 33 |
625 |
0.49 |
| chr18_69349496_69350227 | 0.35 |
Tcf4 |
transcription factor 4 |
917 |
0.69 |
| chr2_134787876_134788027 | 0.35 |
Plcb1 |
phospholipase C, beta 1 |
1421 |
0.42 |
| chr16_50380747_50382000 | 0.35 |
Bbx |
bobby sox HMG box containing |
48539 |
0.18 |
| chr13_93307758_93308718 | 0.34 |
Homer1 |
homer scaffolding protein 1 |
204 |
0.92 |
| chr9_81629834_81629994 | 0.34 |
D430036J16Rik |
RIKEN cDNA D430036J16 gene |
1637 |
0.38 |
| chr19_32866529_32866680 | 0.34 |
Pten |
phosphatase and tensin homolog |
55379 |
0.14 |
| chr8_109250884_109251908 | 0.33 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
1530 |
0.52 |
| chr17_70521708_70522843 | 0.33 |
Dlgap1 |
DLG associated protein 1 |
113 |
0.98 |
| chr2_120244697_120245562 | 0.33 |
Pla2g4e |
phospholipase A2, group IVE |
21 |
0.89 |
| chr2_167233751_167233909 | 0.33 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
6759 |
0.15 |
| chr8_45109829_45110486 | 0.33 |
Gm30504 |
predicted gene, 30504 |
324 |
0.88 |
| chr7_87584791_87585125 | 0.32 |
Grm5 |
glutamate receptor, metabotropic 5 |
560 |
0.85 |
| chr2_92198196_92198596 | 0.31 |
Mir1955 |
microRNA 1955 |
6419 |
0.17 |
| chr11_63412437_63413498 | 0.31 |
B130011K05Rik |
RIKEN cDNA B130011K05 gene |
202 |
0.96 |
| chr7_91092040_91092191 | 0.31 |
Dlg2 |
discs large MAGUK scaffold protein 2 |
1387 |
0.4 |
| chr7_109783136_109783287 | 0.30 |
Nrip3 |
nuclear receptor interacting protein 3 |
1186 |
0.34 |
| chr16_42782105_42782459 | 0.30 |
4932412D23Rik |
RIKEN cDNA 4932412D23 gene |
93305 |
0.08 |
| chr13_28416755_28419194 | 0.30 |
Gm40841 |
predicted gene, 40841 |
1889 |
0.42 |
| chr15_74679054_74679804 | 0.30 |
Arc |
activity regulated cytoskeletal-associated protein |
6859 |
0.1 |
| chr19_16061967_16062187 | 0.29 |
Rpl37-ps1 |
ribosomal protein 37, pseudogene 1 |
36626 |
0.15 |
| chr2_74699510_74699661 | 0.29 |
Gm44463 |
predicted gene, 44463 |
1557 |
0.11 |
| chr4_25798002_25798171 | 0.29 |
Fut9 |
fucosyltransferase 9 |
1769 |
0.36 |
| chr4_153110438_153110637 | 0.29 |
Gm13173 |
predicted gene 13173 |
36406 |
0.21 |
| chr1_35849717_35849997 | 0.29 |
1110002O04Rik |
RIKEN cDNA 1110002O04 gene |
29988 |
0.16 |
| chr2_96319240_96319943 | 0.29 |
Lrrc4c |
leucine rich repeat containing 4C |
1375 |
0.61 |
| chr4_137043562_137044547 | 0.29 |
Zbtb40 |
zinc finger and BTB domain containing 40 |
4641 |
0.19 |
| chr3_34773718_34774341 | 0.28 |
Gm38509 |
predicted gene, 38509 |
1954 |
0.34 |
| chrX_51678321_51678764 | 0.28 |
Hs6st2 |
heparan sulfate 6-O-sulfotransferase 2 |
2023 |
0.41 |
| chr2_59438036_59438472 | 0.28 |
Gm13549 |
predicted gene 13549 |
38473 |
0.13 |
| chr12_54702794_54702945 | 0.28 |
Rpl31-ps17 |
ribosomal protein L31, pseudogene 17 |
1163 |
0.21 |
| chr16_10447447_10447926 | 0.28 |
Tvp23a |
trans-golgi network vesicle protein 23A |
324 |
0.87 |
| chr15_85674019_85674170 | 0.28 |
Lncppara |
long noncoding RNA near Ppara |
20478 |
0.13 |
| chr2_64172800_64172951 | 0.28 |
Fign |
fidgetin |
74837 |
0.13 |
| chr18_54988564_54988904 | 0.27 |
Zfp608 |
zinc finger protein 608 |
1432 |
0.35 |
| chr13_46420548_46420948 | 0.27 |
Rbm24 |
RNA binding motif protein 24 |
197 |
0.96 |
| chr5_14515030_14515887 | 0.27 |
Pclo |
piccolo (presynaptic cytomatrix protein) |
465 |
0.81 |
| chr14_57132708_57133925 | 0.27 |
Gjb6 |
gap junction protein, beta 6 |
35 |
0.97 |
| chr2_145862226_145862910 | 0.27 |
Rin2 |
Ras and Rab interactor 2 |
76 |
0.97 |
| chr18_38928285_38929463 | 0.27 |
Fgf1 |
fibroblast growth factor 1 |
283 |
0.92 |
| chr3_100804632_100805148 | 0.26 |
Vtcn1 |
V-set domain containing T cell activation inhibitor 1 |
20569 |
0.2 |
| chr1_38987342_38987601 | 0.26 |
Pdcl3 |
phosducin-like 3 |
370 |
0.84 |
| chr17_17448568_17449744 | 0.26 |
Lix1 |
limb and CNS expressed 1 |
336 |
0.88 |
| chr1_185193564_185193767 | 0.26 |
Rab3gap2 |
RAB3 GTPase activating protein subunit 2 |
10452 |
0.15 |
| chr10_85389057_85389718 | 0.26 |
Btbd11 |
BTB (POZ) domain containing 11 |
2560 |
0.31 |
| chr5_146921086_146921291 | 0.26 |
Gtf3a |
general transcription factor III A |
27469 |
0.14 |
| chr8_109185308_109186079 | 0.25 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
64173 |
0.13 |
| chr2_167239298_167240893 | 0.25 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
494 |
0.75 |
| chr2_180388160_180389480 | 0.25 |
Mir1a-1 |
microRNA 1a-1 |
228 |
0.89 |
| chr11_42181406_42181557 | 0.25 |
Gabra1 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
623 |
0.84 |
| chr4_123406736_123407057 | 0.25 |
Macf1 |
microtubule-actin crosslinking factor 1 |
5332 |
0.18 |
| chr4_14621039_14622190 | 0.25 |
Slc26a7 |
solute carrier family 26, member 7 |
55 |
0.99 |
| chr6_53817061_53817543 | 0.25 |
Gm16499 |
predicted gene 16499 |
1441 |
0.39 |
| chr9_52680840_52680991 | 0.25 |
Gm1715 |
predicted gene 1715 |
1052 |
0.41 |
| chr10_111100028_111100179 | 0.25 |
Gm48851 |
predicted gene, 48851 |
2768 |
0.22 |
| chr2_65846012_65846807 | 0.25 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
554 |
0.79 |
| chr14_86261599_86262221 | 0.25 |
4930529K09Rik |
RIKEN cDNA 4930529K09 gene |
15907 |
0.12 |
| chrX_7821779_7823061 | 0.25 |
Kcnd1 |
potassium voltage-gated channel, Shal-related family, member 1 |
113 |
0.92 |
| chr7_78569130_78569412 | 0.25 |
Ntrk3 |
neurotrophic tyrosine kinase, receptor, type 3 |
2620 |
0.26 |
| chr17_8388660_8390136 | 0.25 |
Gm5491 |
predicted gene 5491 |
4738 |
0.15 |
| chr19_37907678_37907926 | 0.24 |
Myof |
myoferlin |
1978 |
0.37 |
| chr17_67303669_67304273 | 0.24 |
Ptprm |
protein tyrosine phosphatase, receptor type, M |
50102 |
0.18 |
| chr16_89957880_89958031 | 0.24 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
1848 |
0.47 |
| chr16_20692791_20694082 | 0.24 |
Fam131a |
family with sequence similarity 131, member A |
156 |
0.88 |
| chr2_160367750_160367926 | 0.24 |
Mafb |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
773 |
0.73 |
| chr18_73696846_73697208 | 0.23 |
Smad4 |
SMAD family member 4 |
6753 |
0.16 |
| chr5_21865389_21865953 | 0.23 |
Slc26a5 |
solute carrier family 26, member 5 |
67 |
0.96 |
| chr14_78678524_78678680 | 0.23 |
Gm18368 |
predicted gene, 18368 |
25208 |
0.16 |
| chr4_91376198_91376448 | 0.23 |
Elavl2 |
ELAV like RNA binding protein 1 |
16 |
0.98 |
| chr18_64889550_64889704 | 0.23 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
598 |
0.62 |
| chr2_59438620_59439113 | 0.23 |
Gm13549 |
predicted gene 13549 |
39085 |
0.13 |
| chr14_84437974_84438125 | 0.23 |
Pcdh17 |
protocadherin 17 |
5514 |
0.26 |
| chr9_120098696_120098847 | 0.23 |
Gm47050 |
predicted gene, 47050 |
3108 |
0.11 |
| chr19_46643484_46643643 | 0.23 |
Wbp1l |
WW domain binding protein 1 like |
20162 |
0.11 |
| chr3_31026652_31026803 | 0.23 |
Prkci |
protein kinase C, iota |
4995 |
0.18 |
| chr9_78504198_78504349 | 0.23 |
Eef1a1 |
eukaryotic translation elongation factor 1 alpha 1 |
15122 |
0.11 |
| chr18_65393136_65393441 | 0.23 |
Alpk2 |
alpha-kinase 2 |
606 |
0.6 |
| chr15_44752328_44752563 | 0.23 |
Sybu |
syntabulin (syntaxin-interacting) |
13 |
0.67 |
| chr13_98534689_98535089 | 0.23 |
Gm37425 |
predicted gene, 37425 |
16951 |
0.12 |
| chr9_4794566_4795419 | 0.22 |
Gria4 |
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
527 |
0.88 |
| chr9_117870768_117871285 | 0.22 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
1558 |
0.39 |
| chr10_71127422_71127648 | 0.22 |
Bicc1 |
BicC family RNA binding protein 1 |
19325 |
0.12 |
| chr17_64346137_64346566 | 0.22 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
14435 |
0.23 |
| chr4_53442042_53442694 | 0.22 |
Slc44a1 |
solute carrier family 44, member 1 |
1684 |
0.42 |
| chr19_25343674_25344266 | 0.22 |
Gm34432 |
predicted gene, 34432 |
3233 |
0.31 |
| chr14_91896039_91896190 | 0.22 |
Gm48943 |
predicted gene, 48943 |
219666 |
0.02 |
| chr18_69563797_69564227 | 0.22 |
Tcf4 |
transcription factor 4 |
677 |
0.76 |
| chr4_117913262_117913510 | 0.22 |
Ipo13 |
importin 13 |
1613 |
0.23 |
| chr12_72285979_72286146 | 0.22 |
Rtn1 |
reticulon 1 |
49338 |
0.15 |
| chr19_28165559_28166176 | 0.22 |
Gm35781 |
predicted gene, 35781 |
19564 |
0.23 |
| chr2_60036508_60037747 | 0.22 |
Gm13572 |
predicted gene 13572 |
14 |
0.98 |
| chr1_189910816_189911265 | 0.22 |
Gm38245 |
predicted gene, 38245 |
4919 |
0.19 |
| chr4_47609604_47609755 | 0.22 |
Sec61b |
Sec61 beta subunit |
134731 |
0.05 |
| chr14_31662695_31662846 | 0.22 |
Hacl1 |
2-hydroxyacyl-CoA lyase 1 |
21484 |
0.13 |
| chr10_102515341_102515492 | 0.21 |
Rassf9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
3160 |
0.25 |
| chr5_83352776_83353067 | 0.21 |
Tecrl |
trans-2,3-enoyl-CoA reductase-like |
2035 |
0.43 |
| chr6_54539408_54539559 | 0.21 |
Scrn1 |
secernin 1 |
14963 |
0.17 |
| chr1_163301248_163301399 | 0.21 |
Gm37644 |
predicted gene, 37644 |
7744 |
0.18 |
| chr10_60800688_60800996 | 0.21 |
Gm19972 |
predicted gene, 19972 |
13481 |
0.18 |
| chr17_17910234_17910768 | 0.21 |
Gm7535 |
predicted gene 7535 |
2315 |
0.16 |
| chr8_22508392_22509319 | 0.21 |
Slc20a2 |
solute carrier family 20, member 2 |
2629 |
0.22 |
| chr2_64074730_64075670 | 0.21 |
Fign |
fidgetin |
22788 |
0.29 |
| chr3_102224536_102224803 | 0.21 |
Vangl1 |
VANGL planar cell polarity 1 |
19976 |
0.13 |
| chr2_112591399_112591550 | 0.21 |
Gm44374 |
predicted gene, 44374 |
22556 |
0.16 |
| chr12_117163848_117163999 | 0.21 |
Gm10421 |
predicted gene 10421 |
12272 |
0.28 |
| chr13_32008957_32009151 | 0.21 |
Gm35732 |
predicted gene, 35732 |
37968 |
0.18 |
| chr2_63454866_63455043 | 0.21 |
Gm23503 |
predicted gene, 23503 |
22760 |
0.24 |
| chr8_66700179_66700330 | 0.21 |
Npy1r |
neuropeptide Y receptor Y1 |
2832 |
0.28 |
| chr10_112270073_112271254 | 0.21 |
Kcnc2 |
potassium voltage gated channel, Shaw-related subfamily, member 2 |
458 |
0.85 |
| chr9_44877057_44877237 | 0.21 |
Gm39326 |
predicted gene, 39326 |
744 |
0.44 |
| chr9_66004589_66004841 | 0.21 |
Csnk1g1 |
casein kinase 1, gamma 1 |
3025 |
0.23 |
| chr11_96297868_96298678 | 0.20 |
Hoxb6 |
homeobox B6 |
898 |
0.3 |
| chr9_45370028_45371246 | 0.20 |
Fxyd6 |
FXYD domain-containing ion transport regulator 6 |
195 |
0.91 |
| chr6_8952771_8953600 | 0.20 |
Nxph1 |
neurexophilin 1 |
3509 |
0.38 |
| chr19_14591939_14592995 | 0.20 |
Tle4 |
transducin-like enhancer of split 4 |
3072 |
0.36 |
| chr2_6868272_6868550 | 0.20 |
Celf2 |
CUGBP, Elav-like family member 2 |
3561 |
0.25 |
| chr15_85676712_85677176 | 0.20 |
Lncppara |
long noncoding RNA near Ppara |
23328 |
0.12 |
| chr6_51844040_51844803 | 0.20 |
Skap2 |
src family associated phosphoprotein 2 |
27508 |
0.2 |
| chr10_120721676_120722691 | 0.20 |
Gm37505 |
predicted gene, 37505 |
9632 |
0.13 |
| chr2_10008087_10008238 | 0.20 |
C630004M23Rik |
RIKEN cDNA C630004M23 gene |
39676 |
0.09 |
| chr14_49176028_49176179 | 0.20 |
Naa30 |
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
1796 |
0.31 |
| chr17_44623731_44624125 | 0.20 |
n-R5s27 |
nuclear encoded rRNA 5S 27 |
17215 |
0.22 |
| chr8_121730928_121732115 | 0.20 |
Jph3 |
junctophilin 3 |
954 |
0.49 |
| chr14_64834470_64835221 | 0.20 |
Gm20111 |
predicted gene, 20111 |
4255 |
0.18 |
| chr3_53043747_53045351 | 0.20 |
Gm42901 |
predicted gene 42901 |
2794 |
0.16 |
| chr11_88619965_88620857 | 0.20 |
Msi2 |
musashi RNA-binding protein 2 |
30264 |
0.2 |
| chr19_36118883_36119585 | 0.20 |
Ankrd1 |
ankyrin repeat domain 1 (cardiac muscle) |
676 |
0.71 |
| chr6_86032524_86032675 | 0.20 |
Add2 |
adducin 2 (beta) |
3852 |
0.13 |
| chr17_22339851_22340002 | 0.20 |
Gm49951 |
predicted gene, 49951 |
6932 |
0.15 |
| chr18_38598277_38598596 | 0.20 |
Spry4 |
sprouty RTK signaling antagonist 4 |
2979 |
0.2 |
| chr17_79451074_79451933 | 0.19 |
Cdc42ep3 |
CDC42 effector protein (Rho GTPase binding) 3 |
96412 |
0.07 |
| chr4_5645693_5646152 | 0.19 |
Fam110b |
family with sequence similarity 110, member B |
1687 |
0.36 |
| chr2_137356841_137357676 | 0.19 |
Gm14055 |
predicted gene 14055 |
68755 |
0.13 |
| chr2_9869939_9870090 | 0.19 |
Gata3 |
GATA binding protein 3 |
3658 |
0.15 |
| chr3_87767082_87768667 | 0.19 |
Pear1 |
platelet endothelial aggregation receptor 1 |
1061 |
0.43 |
| chr16_72905751_72905902 | 0.19 |
Robo1 |
roundabout guidance receptor 1 |
70523 |
0.14 |
| chrX_51678767_51679193 | 0.19 |
Hs6st2 |
heparan sulfate 6-O-sulfotransferase 2 |
1585 |
0.48 |
| chr16_46493311_46493865 | 0.19 |
Nectin3 |
nectin cell adhesion molecule 3 |
3184 |
0.34 |
| chr4_25796073_25796399 | 0.19 |
Fut9 |
fucosyltransferase 9 |
3619 |
0.24 |
| chr14_110753127_110753447 | 0.19 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
1862 |
0.32 |
| chr7_54835204_54836499 | 0.19 |
Luzp2 |
leucine zipper protein 2 |
236 |
0.94 |
| chr7_122044795_122044946 | 0.19 |
Ears2 |
glutamyl-tRNA synthetase 2, mitochondrial |
22141 |
0.1 |
| chrX_159259045_159259196 | 0.19 |
Rps6ka3 |
ribosomal protein S6 kinase polypeptide 3 |
2994 |
0.34 |
| chr5_25098295_25098663 | 0.19 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
2163 |
0.24 |
| chr4_46202294_46203178 | 0.19 |
Xpa |
xeroderma pigmentosum, complementation group A |
6425 |
0.17 |
| chr8_84770692_84772567 | 0.19 |
Nfix |
nuclear factor I/X |
1767 |
0.22 |
| chr6_138365294_138365445 | 0.19 |
Lmo3 |
LIM domain only 3 |
56083 |
0.13 |
| chr5_75247144_75247638 | 0.19 |
Gm42799 |
predicted gene 42799 |
948 |
0.57 |
| chr3_79576778_79577000 | 0.19 |
Gm35067 |
predicted gene, 35067 |
5699 |
0.12 |
| chr17_22360731_22361353 | 0.19 |
Zfp944 |
zinc finger protein 944 |
358 |
0.51 |
| chr18_41860653_41861741 | 0.19 |
Gm50410 |
predicted gene, 50410 |
13637 |
0.22 |
| chr9_76014472_76015728 | 0.18 |
Hmgcll1 |
3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 |
45 |
0.55 |
| chr1_83517764_83517915 | 0.18 |
Gm37989 |
predicted gene, 37989 |
90441 |
0.08 |
| chr4_23829464_23829682 | 0.18 |
Gm28448 |
predicted gene 28448 |
104381 |
0.08 |
| chr10_24312237_24312511 | 0.18 |
Moxd1 |
monooxygenase, DBH-like 1 |
88829 |
0.07 |
| chr9_101401666_101401952 | 0.18 |
Gm24638 |
predicted gene, 24638 |
8699 |
0.18 |
| chr19_36115709_36115956 | 0.18 |
Ankrd1 |
ankyrin repeat domain 1 (cardiac muscle) |
4078 |
0.23 |
| chr6_92532985_92533460 | 0.18 |
Prickle2 |
prickle planar cell polarity protein 2 |
1639 |
0.46 |
| chr6_116056244_116057282 | 0.18 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
192 |
0.94 |
| chr6_48626698_48627876 | 0.18 |
AI854703 |
expressed sequence AI854703 |
6 |
0.93 |
| chr18_54984578_54984982 | 0.18 |
Zfp608 |
zinc finger protein 608 |
5386 |
0.22 |
| chr9_45404342_45404493 | 0.18 |
Fxyd2 |
FXYD domain-containing ion transport regulator 2 |
441 |
0.75 |
| chr14_79450824_79451101 | 0.18 |
Kbtbd6 |
kelch repeat and BTB (POZ) domain containing 6 |
873 |
0.51 |
| chr2_74694827_74695683 | 0.18 |
Gm14396 |
predicted gene 14396 |
265 |
0.7 |
| chr13_59431296_59431956 | 0.18 |
4930455M05Rik |
RIKEN cDNA 4930455M05 gene |
12229 |
0.14 |
| chr11_81148614_81148842 | 0.18 |
Asic2 |
acid-sensing (proton-gated) ion channel 2 |
4593 |
0.3 |
| chr6_128596921_128597883 | 0.18 |
Gm44009 |
predicted gene, 44009 |
9377 |
0.09 |
| chr6_32584464_32585789 | 0.18 |
Plxna4 |
plexin A4 |
3066 |
0.3 |
| chr1_23490467_23490664 | 0.17 |
Gm7784 |
predicted gene 7784 |
11026 |
0.24 |
| chr8_50120264_50120415 | 0.17 |
Gm45323 |
predicted gene 45323 |
106955 |
0.07 |
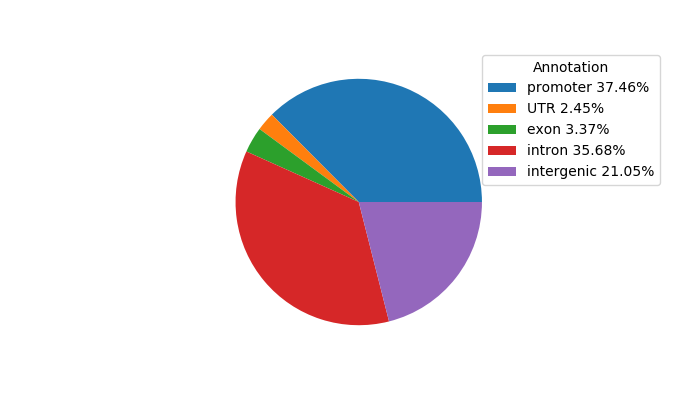
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.6 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.3 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.0 | 0.0 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.2 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0042160 | lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.0 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0003133 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0072262 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.0 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.0 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:1903286 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.0 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.0 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0009445 | putrescine metabolic process(GO:0009445) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0034824 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0004118 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME CELL CELL COMMUNICATION | Genes involved in Cell-Cell communication |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |