Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
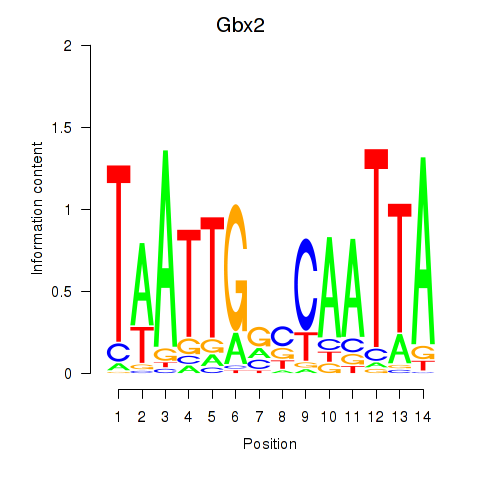
Results for Gbx2
Z-value: 0.71
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSMUSG00000034486.7 | gastrulation brain homeobox 2 |
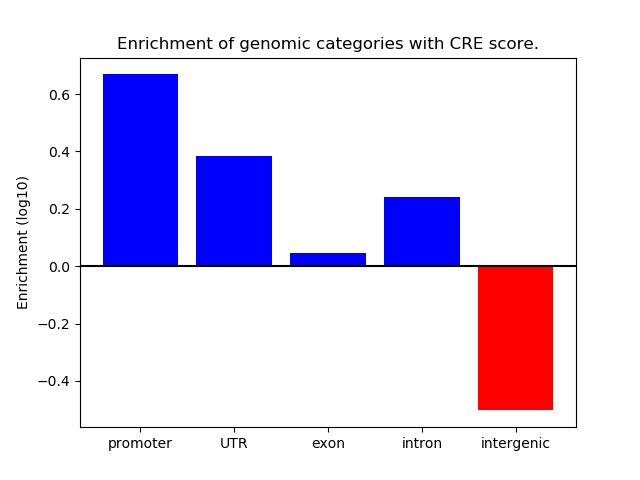
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_89939935_89940647 | Gbx2 | 9112 | 0.195560 | -0.31 | 1.7e-02 | Click! |
| chr1_89921735_89923880 | Gbx2 | 6378 | 0.208198 | -0.28 | 2.7e-02 | Click! |
| chr1_89942989_89943140 | Gbx2 | 11885 | 0.189360 | -0.24 | 6.7e-02 | Click! |
| chr1_89941432_89942704 | Gbx2 | 10889 | 0.191430 | -0.23 | 8.0e-02 | Click! |
| chr1_89939435_89939586 | Gbx2 | 8331 | 0.197710 | -0.22 | 9.7e-02 | Click! |
Activity of the Gbx2 motif across conditions
Conditions sorted by the z-value of the Gbx2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
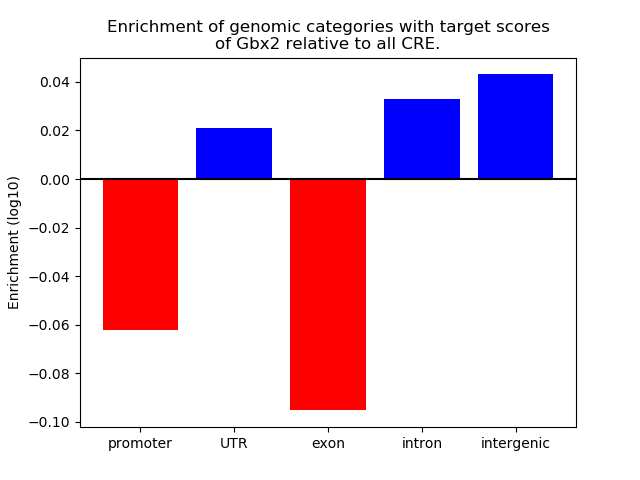
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_21484558_21485279 | 1.53 |
Agtr2 |
angiotensin II receptor, type 2 |
196 |
0.94 |
| chr16_88599651_88600158 | 1.31 |
Gm49688 |
predicted gene, 49688 |
8766 |
0.11 |
| chr2_74696421_74697223 | 1.18 |
Gm28793 |
predicted gene 28793 |
764 |
0.23 |
| chr7_49907312_49908741 | 1.17 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
2120 |
0.4 |
| chrX_58035394_58035659 | 1.08 |
Zic3 |
zinc finger protein of the cerebellum 3 |
4516 |
0.29 |
| chr12_77791074_77791423 | 1.08 |
Gm24070 |
predicted gene, 24070 |
23561 |
0.2 |
| chr12_41483283_41485192 | 1.05 |
Lrrn3 |
leucine rich repeat protein 3, neuronal |
2194 |
0.36 |
| chr16_74407688_74408034 | 1.04 |
Robo2 |
roundabout guidance receptor 2 |
3051 |
0.35 |
| chr9_74869680_74869831 | 1.02 |
Onecut1 |
one cut domain, family member 1 |
3271 |
0.2 |
| chr6_52216788_52218128 | 1.01 |
Hoxa7 |
homeobox A7 |
6 |
0.91 |
| chr14_84450287_84451113 | 1.00 |
Pcdh17 |
protocadherin 17 |
2193 |
0.37 |
| chr16_88561792_88563295 | 0.98 |
Cldn8 |
claudin 8 |
640 |
0.64 |
| chr3_20365498_20365649 | 0.88 |
Agtr1b |
angiotensin II receptor, type 1b |
1551 |
0.38 |
| chr2_11884018_11884169 | 0.86 |
Gm34768 |
predicted gene, 34768 |
15989 |
0.23 |
| chr16_81203532_81203683 | 0.86 |
Ncam2 |
neural cell adhesion molecule 2 |
2850 |
0.36 |
| chr10_126169548_126169835 | 0.85 |
Gm4510 |
predicted gene 4510 |
16632 |
0.26 |
| chr5_131585523_131586662 | 0.85 |
Gm27266 |
predicted gene, 27266 |
8105 |
0.13 |
| chr14_122480308_122481080 | 0.84 |
Zic2 |
zinc finger protein of the cerebellum 2 |
2594 |
0.16 |
| chr3_5224377_5225076 | 0.83 |
Zfhx4 |
zinc finger homeodomain 4 |
3221 |
0.24 |
| chr6_52211753_52213405 | 0.81 |
Hoxa3 |
homeobox A3 |
536 |
0.36 |
| chr4_62966572_62967137 | 0.81 |
Zfp618 |
zinc finger protein 618 |
1280 |
0.42 |
| chr4_154630838_154632339 | 0.80 |
Prdm16 |
PR domain containing 16 |
5209 |
0.14 |
| chr4_22485441_22485749 | 0.77 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
2771 |
0.23 |
| chr12_40449188_40449922 | 0.77 |
Dock4 |
dedicator of cytokinesis 4 |
3219 |
0.27 |
| chr12_51000370_51001072 | 0.76 |
Gm40421 |
predicted gene, 40421 |
4152 |
0.24 |
| chr8_114707514_114708076 | 0.76 |
Gm16118 |
predicted gene 16118 |
211 |
0.95 |
| chr10_109008310_109009456 | 0.76 |
Syt1 |
synaptotagmin I |
217 |
0.96 |
| chr5_43240853_43241007 | 0.75 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
3615 |
0.18 |
| chr9_41578714_41578969 | 0.74 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
653 |
0.53 |
| chr9_103112824_103112975 | 0.74 |
Rab6b |
RAB6B, member RAS oncogene family |
1092 |
0.45 |
| chr13_44026235_44027511 | 0.74 |
Gm33489 |
predicted gene, 33489 |
8351 |
0.22 |
| chr14_99301075_99301422 | 0.74 |
Klf5 |
Kruppel-like factor 5 |
2094 |
0.28 |
| chr15_103058659_103059955 | 0.73 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
1 |
0.95 |
| chr1_115688816_115689173 | 0.73 |
Cntnap5a |
contactin associated protein-like 5A |
4238 |
0.28 |
| chr2_22738184_22738444 | 0.72 |
1700092C17Rik |
RIKEN cDNA 1700092C17 gene |
17670 |
0.13 |
| chr2_155590226_155590377 | 0.72 |
Gssos2 |
glutathione synthase, opposite strand 2 |
830 |
0.34 |
| chr2_116051067_116051481 | 0.72 |
Meis2 |
Meis homeobox 2 |
1594 |
0.37 |
| chr15_102948559_102948937 | 0.71 |
Hotair |
HOX transcript antisense RNA (non-protein coding) |
1018 |
0.31 |
| chr13_63278777_63280117 | 0.71 |
Gm47602 |
predicted gene, 47602 |
223 |
0.69 |
| chr1_124043338_124043489 | 0.71 |
Dpp10 |
dipeptidylpeptidase 10 |
1750 |
0.52 |
| chr3_17326564_17327148 | 0.71 |
Gm30340 |
predicted gene, 30340 |
7569 |
0.24 |
| chr15_99056560_99057587 | 0.70 |
Prph |
peripherin |
1103 |
0.3 |
| chr7_62416059_62417205 | 0.69 |
Mkrn3 |
makorin, ring finger protein, 3 |
3507 |
0.2 |
| chr3_38894285_38895428 | 0.67 |
Fat4 |
FAT atypical cadherin 4 |
3914 |
0.27 |
| chr17_91090702_91091377 | 0.67 |
Nrxn1 |
neurexin I |
1694 |
0.28 |
| chr12_108424381_108424697 | 0.67 |
Eml1 |
echinoderm microtubule associated protein like 1 |
1461 |
0.41 |
| chr16_77594640_77595970 | 0.67 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
403 |
0.71 |
| chr15_32923267_32923822 | 0.67 |
Sdc2 |
syndecan 2 |
2821 |
0.38 |
| chr10_72299004_72299155 | 0.67 |
Gm9923 |
predicted pseudogene 9923 |
10242 |
0.31 |
| chr6_52252411_52254268 | 0.67 |
9530018H14Rik |
RIKEN cDNA 9530018H14 gene |
4121 |
0.07 |
| chr5_144483749_144483900 | 0.65 |
Nptx2 |
neuronal pentraxin 2 |
62078 |
0.11 |
| chr3_55778948_55779699 | 0.64 |
Mab21l1 |
mab-21-like 1 |
3187 |
0.25 |
| chr13_47566403_47566695 | 0.64 |
4930471G24Rik |
RIKEN cDNA 4930471G24 gene |
101001 |
0.08 |
| chr9_30918491_30920140 | 0.64 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
1635 |
0.4 |
| chr19_20009817_20010437 | 0.64 |
Gm22684 |
predicted gene, 22684 |
23508 |
0.22 |
| chr17_22760154_22760305 | 0.64 |
Gm5493 |
predicted gene 5493 |
20284 |
0.13 |
| chr9_91359026_91359334 | 0.64 |
Zic4 |
zinc finger protein of the cerebellum 4 |
3233 |
0.14 |
| chr8_125914178_125914599 | 0.64 |
Map3k21 |
mitogen-activated protein kinase kinase kinase 21 |
3938 |
0.22 |
| chr4_118288250_118288401 | 0.64 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
2949 |
0.24 |
| chr4_33926104_33927188 | 0.63 |
Cnr1 |
cannabinoid receptor 1 (brain) |
444 |
0.88 |
| chr12_29529828_29531185 | 0.63 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr8_79372300_79372589 | 0.62 |
Smad1 |
SMAD family member 1 |
15766 |
0.17 |
| chr1_182823768_182824187 | 0.62 |
Susd4 |
sushi domain containing 4 |
59082 |
0.11 |
| chr6_52224853_52226609 | 0.62 |
Hoxa9 |
homeobox A9 |
458 |
0.51 |
| chr10_97609680_97609831 | 0.62 |
Kera |
keratocan |
2876 |
0.22 |
| chr1_136228373_136230942 | 0.62 |
Inava |
innate immunity activator |
362 |
0.76 |
| chr1_74892200_74893701 | 0.62 |
Cryba2 |
crystallin, beta A2 |
58 |
0.95 |
| chr5_53052069_53052288 | 0.61 |
Slc34a2 |
solute carrier family 34 (sodium phosphate), member 2 |
2825 |
0.22 |
| chr7_29496118_29496269 | 0.61 |
Sipa1l3 |
signal-induced proliferation-associated 1 like 3 |
9259 |
0.16 |
| chrX_7739896_7741560 | 0.61 |
Ccdc120 |
coiled-coil domain containing 120 |
497 |
0.57 |
| chr16_43235566_43236569 | 0.60 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
28 |
0.98 |
| chr3_33269889_33270040 | 0.60 |
Gm27974 |
predicted gene, 27974 |
9144 |
0.2 |
| chr16_67610470_67610655 | 0.60 |
Cadm2 |
cell adhesion molecule 2 |
9931 |
0.25 |
| chr2_22626092_22626243 | 0.59 |
Gad2 |
glutamic acid decarboxylase 2 |
2863 |
0.19 |
| chr15_103009345_103011570 | 0.59 |
Hoxc6 |
homeobox C6 |
884 |
0.35 |
| chr7_79507974_79509311 | 0.58 |
A330074H02Rik |
RIKEN cDNA A330074H02 gene |
1720 |
0.18 |
| chr3_29957475_29957868 | 0.58 |
Gm37281 |
predicted gene, 37281 |
31135 |
0.15 |
| chr8_45688068_45688642 | 0.58 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
18 |
0.98 |
| chr15_25408835_25409622 | 0.58 |
Basp1 |
brain abundant, membrane attached signal protein 1 |
4470 |
0.17 |
| chr11_96282910_96285155 | 0.58 |
Hoxb8 |
homeobox B8 |
1307 |
0.19 |
| chr2_28910187_28910661 | 0.57 |
Barhl1 |
BarH like homeobox 1 |
2071 |
0.29 |
| chr1_78186833_78187893 | 0.57 |
Pax3 |
paired box 3 |
9475 |
0.23 |
| chr15_32780266_32781493 | 0.57 |
Gm32618 |
predicted gene, 32618 |
1975 |
0.38 |
| chr15_103105552_103105797 | 0.56 |
Gm28265 |
predicted gene 28265 |
1072 |
0.32 |
| chr11_120242307_120244045 | 0.56 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
6477 |
0.1 |
| chr6_107711293_107711898 | 0.55 |
4933431M02Rik |
RIKEN cDNA 4933431M02 gene |
83821 |
0.1 |
| chr6_42707641_42707961 | 0.55 |
Tcaf1 |
TRPM8 channel-associated factor 1 |
2270 |
0.16 |
| chr5_7342956_7343453 | 0.55 |
Zfp804b |
zinc finger protein 804B |
1552 |
0.41 |
| chr18_69595338_69595620 | 0.54 |
Tcf4 |
transcription factor 4 |
1297 |
0.56 |
| chr5_84413029_84413589 | 0.54 |
Epha5 |
Eph receptor A5 |
3497 |
0.31 |
| chr14_104030791_104030942 | 0.54 |
4930432J09Rik |
RIKEN cDNA 4930432J09 gene |
3724 |
0.34 |
| chr8_12915306_12916734 | 0.54 |
Mcf2l |
mcf.2 transforming sequence-like |
45 |
0.76 |
| chr11_86605282_86605433 | 0.54 |
Vmp1 |
vacuole membrane protein 1 |
3062 |
0.2 |
| chr7_48961270_48961876 | 0.54 |
Nav2 |
neuron navigator 2 |
2476 |
0.25 |
| chr12_73997086_73997237 | 0.53 |
Syt16 |
synaptotagmin XVI |
500 |
0.77 |
| chr5_47796679_47796830 | 0.53 |
Gm3010 |
predicted gene 3010 |
96909 |
0.08 |
| chr9_118199526_118199677 | 0.53 |
Gm17399 |
predicted gene, 17399 |
49370 |
0.14 |
| chr17_90452047_90452824 | 0.53 |
Nrxn1 |
neurexin I |
2387 |
0.31 |
| chr9_52500605_52500756 | 0.53 |
Gm49389 |
predicted gene, 49389 |
96381 |
0.07 |
| chr2_167540595_167541629 | 0.52 |
Snai1 |
snail family zinc finger 1 |
2917 |
0.16 |
| chr3_45204718_45204869 | 0.52 |
Gm27989 |
predicted gene, 27989 |
44098 |
0.15 |
| chr3_95163200_95164363 | 0.52 |
Sema6c |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
525 |
0.54 |
| chr6_144202688_144204608 | 0.52 |
Sox5 |
SRY (sex determining region Y)-box 5 |
409 |
0.92 |
| chr12_12937582_12937733 | 0.51 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
2959 |
0.18 |
| chr5_114569026_114570120 | 0.51 |
Fam222a |
family with sequence similarity 222, member A |
1556 |
0.35 |
| chr19_41739688_41740472 | 0.51 |
Slit1 |
slit guidance ligand 1 |
3406 |
0.26 |
| chr1_157798825_157799345 | 0.51 |
Gm38256 |
predicted gene, 38256 |
141300 |
0.04 |
| chr7_62208910_62209522 | 0.51 |
Gm9801 |
predicted gene 9801 |
263 |
0.94 |
| chr2_165596319_165596667 | 0.51 |
Eya2 |
EYA transcriptional coactivator and phosphatase 2 |
1461 |
0.39 |
| chr1_62704948_62705819 | 0.51 |
Nrp2 |
neuropilin 2 |
1681 |
0.35 |
| chr6_52196485_52198020 | 0.51 |
Hoxaas3 |
Hoxa cluster antisense RNA 3 |
3872 |
0.06 |
| chr17_64206242_64206631 | 0.50 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
106595 |
0.07 |
| chr15_95525022_95525792 | 0.50 |
Nell2 |
NEL-like 2 |
2771 |
0.35 |
| chr8_100228228_100228379 | 0.50 |
Gm5742 |
predicted gene 5742 |
168187 |
0.04 |
| chr2_153440130_153440281 | 0.50 |
Nol4l |
nucleolar protein 4-like |
4274 |
0.2 |
| chr13_51408385_51410292 | 0.50 |
S1pr3 |
sphingosine-1-phosphate receptor 3 |
699 |
0.69 |
| chr8_89288677_89288828 | 0.50 |
Gm5356 |
predicted pseudogene 5356 |
101192 |
0.08 |
| chr5_147303898_147304049 | 0.50 |
Cdx2 |
caudal type homeobox 2 |
3297 |
0.13 |
| chr6_37645617_37646179 | 0.50 |
Ybx1-ps2 |
Y box protein 1, pseudogene 2 |
41297 |
0.18 |
| chr8_48136896_48137754 | 0.49 |
Dctd |
dCMP deaminase |
27114 |
0.2 |
| chr11_95597886_95598472 | 0.49 |
Ngfr |
nerve growth factor receptor (TNFR superfamily, member 16) |
10444 |
0.16 |
| chr16_72811501_72811652 | 0.49 |
Robo1 |
roundabout guidance receptor 1 |
148372 |
0.05 |
| chr14_118435860_118436192 | 0.49 |
Gm5672 |
predicted gene 5672 |
59832 |
0.09 |
| chr17_68836792_68837954 | 0.49 |
Gm38593 |
predicted gene, 38593 |
147 |
0.62 |
| chr9_122597077_122597558 | 0.49 |
9530059O14Rik |
RIKEN cDNA 9530059O14 gene |
24815 |
0.12 |
| chr1_41605412_41605563 | 0.48 |
Gm28634 |
predicted gene 28634 |
75944 |
0.12 |
| chr1_138837882_138838896 | 0.48 |
Lhx9 |
LIM homeobox protein 9 |
4040 |
0.18 |
| chr18_69349496_69350227 | 0.48 |
Tcf4 |
transcription factor 4 |
917 |
0.69 |
| chr4_126150157_126151254 | 0.47 |
Eva1b |
eva-1 homolog B (C. elegans) |
2173 |
0.18 |
| chr5_120433178_120434996 | 0.47 |
Gm27199 |
predicted gene 27199 |
2320 |
0.19 |
| chr12_59129998_59130231 | 0.47 |
Mia2 |
MIA SH3 domain ER export factor 2 |
263 |
0.88 |
| chr10_92161472_92161916 | 0.46 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
1067 |
0.55 |
| chr6_52176961_52178435 | 0.46 |
5730596B20Rik |
RIKEN cDNA 5730596B20 gene |
200 |
0.79 |
| chr9_63139436_63140636 | 0.46 |
Gm25064 |
predicted gene, 25064 |
6753 |
0.18 |
| chr12_84493482_84493633 | 0.46 |
Lin52 |
lin-52 homolog (C. elegans) |
36161 |
0.11 |
| chr13_83713464_83713615 | 0.46 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
7842 |
0.14 |
| chr3_26331036_26332158 | 0.46 |
Nlgn1 |
neuroligin 1 |
273 |
0.58 |
| chr7_5056231_5057716 | 0.46 |
Gm15510 |
predicted gene 15510 |
53 |
0.48 |
| chr4_150270799_150270951 | 0.45 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
10771 |
0.14 |
| chr2_169178799_169179344 | 0.45 |
9430093N23Rik |
RIKEN cDNA 9430093N23 gene |
55920 |
0.13 |
| chr5_67466735_67467291 | 0.45 |
C330024D21Rik |
RIKEN cDNA C330024D21 gene |
3110 |
0.19 |
| chr12_52911080_52911262 | 0.45 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
115335 |
0.06 |
| chr3_17796058_17796354 | 0.45 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
462 |
0.52 |
| chr1_62564755_62565185 | 0.45 |
Gm29084 |
predicted gene 29084 |
29028 |
0.18 |
| chr2_52720638_52720837 | 0.45 |
Gm13548 |
predicted gene 13548 |
18718 |
0.18 |
| chr11_78170207_78170358 | 0.44 |
Nek8 |
NIMA (never in mitosis gene a)-related expressed kinase 8 |
454 |
0.56 |
| chr1_174920917_174922100 | 0.44 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
311 |
0.94 |
| chr8_35883104_35883618 | 0.44 |
5430403N17Rik |
RIKEN cDNA 5430403N17 gene |
404 |
0.86 |
| chr1_138835333_138836550 | 0.44 |
Lhx9 |
LIM homeobox protein 9 |
2282 |
0.24 |
| chr7_28203492_28203757 | 0.44 |
Gm45011 |
predicted gene 45011 |
22734 |
0.08 |
| chr7_133608062_133608429 | 0.44 |
Gm45671 |
predicted gene 45671 |
19 |
0.96 |
| chr5_33723808_33724192 | 0.44 |
Fgfr3 |
fibroblast growth factor receptor 3 |
228 |
0.59 |
| chr19_60148618_60149048 | 0.44 |
E330013P04Rik |
RIKEN cDNA E330013P04 gene |
2461 |
0.3 |
| chr8_61309431_61310318 | 0.43 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
21002 |
0.18 |
| chr13_67039116_67039380 | 0.43 |
4933433G19Rik |
RIKEN cDNA 4933433G19 gene |
6840 |
0.1 |
| chr10_111247804_111248910 | 0.43 |
Osbpl8 |
oxysterol binding protein-like 8 |
289 |
0.91 |
| chr3_148984296_148985724 | 0.43 |
Gm43573 |
predicted gene 43573 |
4142 |
0.2 |
| chr19_58076835_58077494 | 0.43 |
Gm50287 |
predicted gene, 50287 |
18867 |
0.22 |
| chr2_73777255_73777445 | 0.43 |
Chn1 |
chimerin 1 |
2004 |
0.35 |
| chr3_110139298_110139994 | 0.43 |
Ntng1 |
netrin G1 |
3358 |
0.33 |
| chr11_36674002_36674437 | 0.43 |
Tenm2 |
teneurin transmembrane protein 2 |
3526 |
0.35 |
| chrX_12469610_12469761 | 0.43 |
Rpl30-ps11 |
ribosomal protein L30, pseudogene 11 |
12455 |
0.25 |
| chr8_12428084_12429088 | 0.43 |
Gm25239 |
predicted gene, 25239 |
32183 |
0.11 |
| chr12_46814495_46815083 | 0.43 |
Nova1 |
NOVA alternative splicing regulator 1 |
2171 |
0.31 |
| chr13_72288993_72289292 | 0.43 |
Gm4052 |
predicted gene 4052 |
61079 |
0.13 |
| chr14_67241509_67241827 | 0.43 |
Ebf2 |
early B cell factor 2 |
7024 |
0.17 |
| chr15_25415436_25415919 | 0.43 |
Gm48957 |
predicted gene, 48957 |
614 |
0.58 |
| chr3_138280115_138280773 | 0.42 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
2793 |
0.17 |
| chr14_104640336_104640571 | 0.42 |
D130009I18Rik |
RIKEN cDNA D130009I18 gene |
1309 |
0.47 |
| chr4_153480513_153480981 | 0.42 |
Ajap1 |
adherens junction associated protein 1 |
1432 |
0.58 |
| chr11_22284673_22285000 | 0.42 |
Ehbp1 |
EH domain binding protein 1 |
1002 |
0.67 |
| chr2_77701567_77703605 | 0.42 |
Zfp385b |
zinc finger protein 385B |
686 |
0.8 |
| chr16_45098155_45099397 | 0.42 |
Ccdc80 |
coiled-coil domain containing 80 |
4723 |
0.24 |
| chr2_31638722_31641540 | 0.42 |
Prdm12 |
PR domain containing 12 |
94 |
0.84 |
| chr14_122479539_122479995 | 0.42 |
Zic2 |
zinc finger protein of the cerebellum 2 |
1667 |
0.23 |
| chr9_78175193_78175344 | 0.42 |
C920006O11Rik |
RIKEN cDNA C920006O11 gene |
646 |
0.58 |
| chr15_13067157_13068365 | 0.41 |
Gm24664 |
predicted gene, 24664 |
86932 |
0.08 |
| chr11_103339490_103340754 | 0.41 |
Arhgap27 |
Rho GTPase activating protein 27 |
403 |
0.77 |
| chr9_45433206_45433653 | 0.41 |
4833428L15Rik |
RIKEN cDNA 4833428L15 gene |
1699 |
0.26 |
| chr13_58450895_58451149 | 0.41 |
Gm47918 |
predicted gene, 47918 |
8001 |
0.16 |
| chr18_40260537_40261051 | 0.41 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
1991 |
0.29 |
| chrX_94637542_94637693 | 0.41 |
Gspt2 |
G1 to S phase transition 2 |
1541 |
0.3 |
| chr12_50121867_50122018 | 0.41 |
Gm40418 |
predicted gene, 40418 |
1633 |
0.56 |
| chr15_78116860_78117881 | 0.41 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
2336 |
0.22 |
| chr18_13967357_13968027 | 0.41 |
Zfp521 |
zinc finger protein 521 |
4028 |
0.33 |
| chr2_116053626_116054503 | 0.40 |
Meis2 |
Meis homeobox 2 |
4384 |
0.2 |
| chr15_64916306_64916931 | 0.40 |
Adcy8 |
adenylate cyclase 8 |
5653 |
0.28 |
| chr13_85067537_85067966 | 0.40 |
Gm47745 |
predicted gene, 47745 |
26584 |
0.17 |
| chr2_33713186_33713886 | 0.40 |
9430024E24Rik |
RIKEN cDNA 9430024E24 gene |
5363 |
0.21 |
| chr19_16437610_16437973 | 0.40 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
1509 |
0.37 |
| chr19_28500495_28500646 | 0.40 |
Glis3 |
GLIS family zinc finger 3 |
39850 |
0.19 |
| chr12_30370291_30370442 | 0.40 |
Sntg2 |
syntrophin, gamma 2 |
2899 |
0.33 |
| chr2_121803106_121803474 | 0.40 |
Frmd5 |
FERM domain containing 5 |
3582 |
0.23 |
| chr7_81341709_81341934 | 0.40 |
Rps17 |
ribosomal protein S17 |
2710 |
0.22 |
| chr18_21654600_21655235 | 0.40 |
Klhl14 |
kelch-like 14 |
199 |
0.95 |
| chr1_137088325_137088476 | 0.40 |
Gm23763 |
predicted gene, 23763 |
98130 |
0.07 |
| chr5_5431980_5432131 | 0.40 |
Cdk14 |
cyclin-dependent kinase 14 |
11743 |
0.19 |
| chr5_117841452_117842996 | 0.40 |
Nos1 |
nitric oxide synthase 1, neuronal |
38 |
0.98 |
| chr16_42337545_42337696 | 0.40 |
Gap43 |
growth associated protein 43 |
3031 |
0.33 |
| chr14_89793420_89793784 | 0.40 |
Gm25415 |
predicted gene, 25415 |
86797 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.2 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.6 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.2 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 0.5 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.2 | 0.9 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 0.4 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.4 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.6 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.3 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.4 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.3 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.3 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.2 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.1 | 0.3 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.1 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.1 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 0.2 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.0 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.2 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.3 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.3 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.2 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0035793 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0021855 | hypothalamus cell migration(GO:0021855) |
| 0.0 | 0.0 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0032803 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.1 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.0 | GO:0072203 | cell proliferation involved in metanephros development(GO:0072203) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.0 | 0.0 | GO:0045636 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.0 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.0 | GO:0021604 | cranial nerve structural organization(GO:0021604) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0035247 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:0072172 | mesonephric tubule formation(GO:0072172) |
| 0.0 | 0.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.2 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.0 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 0.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0052830 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |