Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
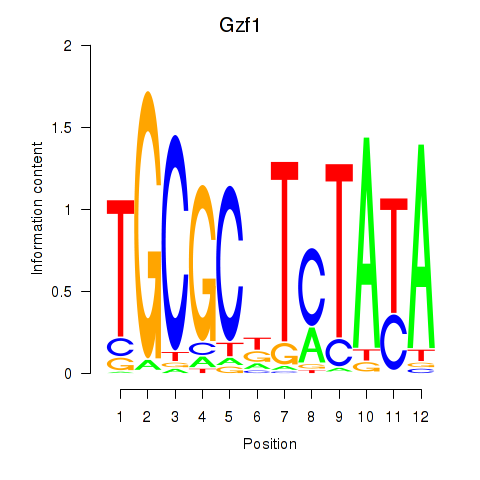
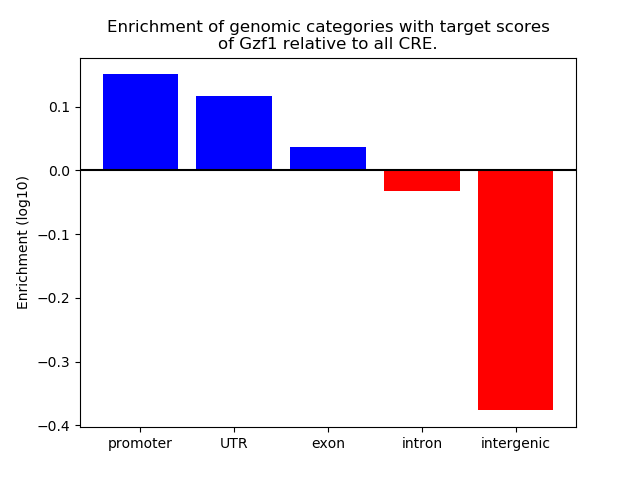
Results for Gzf1
Z-value: 0.33
Transcription factors associated with Gzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gzf1
|
ENSMUSG00000027439.9 | GDNF-inducible zinc finger protein 1 |
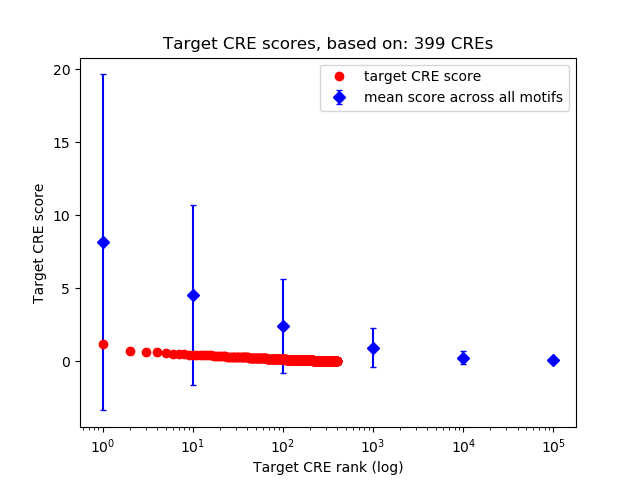
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_148680576_148680950 | Gzf1 | 260 | 0.897015 | 0.38 | 2.6e-03 | Click! |
| chr2_148678935_148679497 | Gzf1 | 1807 | 0.288201 | -0.33 | 9.9e-03 | Click! |
| chr2_148678060_148678476 | Gzf1 | 2755 | 0.213132 | -0.27 | 3.8e-02 | Click! |
| chr2_148682624_148682775 | Gzf1 | 1488 | 0.339850 | 0.14 | 2.9e-01 | Click! |
| chr2_148677237_148678042 | Gzf1 | 3384 | 0.190726 | -0.07 | 5.8e-01 | Click! |
Activity of the Gzf1 motif across conditions
Conditions sorted by the z-value of the Gzf1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
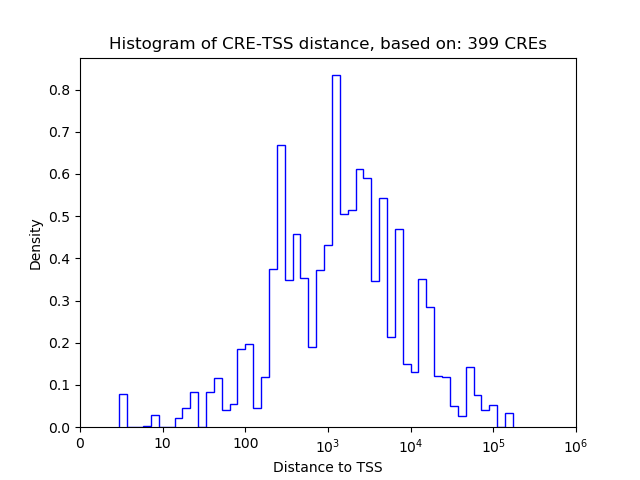
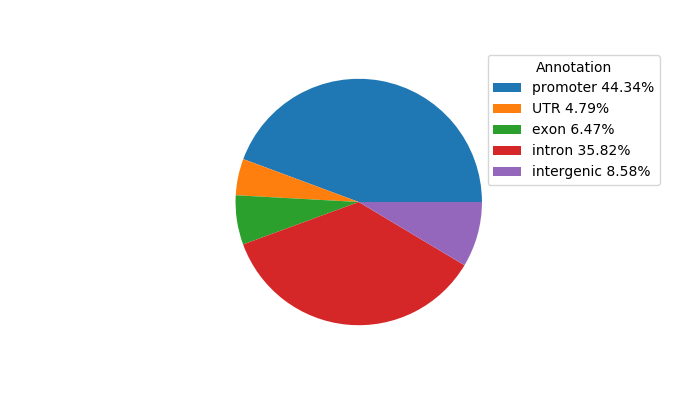
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_80258361_80258974 | 1.18 |
Slc66a2 |
solute carrier family 66 member 2 |
2349 |
0.18 |
| chr1_9776842_9777028 | 0.72 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
6843 |
0.14 |
| chr17_25089311_25089630 | 0.62 |
Ift140 |
intraflagellar transport 140 |
1201 |
0.31 |
| chr1_40430303_40430529 | 0.61 |
Il1rl1 |
interleukin 1 receptor-like 1 |
846 |
0.63 |
| chr9_43234131_43234830 | 0.54 |
Oaf |
out at first homolog |
5431 |
0.16 |
| chr15_75906413_75907979 | 0.51 |
Eef1d |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
1833 |
0.15 |
| chr12_111445027_111446639 | 0.51 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
251 |
0.87 |
| chr4_132396755_132396912 | 0.47 |
Phactr4 |
phosphatase and actin regulator 4 |
1366 |
0.24 |
| chr16_91466045_91466196 | 0.45 |
Gm49626 |
predicted gene, 49626 |
995 |
0.24 |
| chr19_60871718_60871869 | 0.44 |
Prdx3 |
peroxiredoxin 3 |
2763 |
0.19 |
| chr8_13108983_13109168 | 0.43 |
Cul4a |
cullin 4A |
3139 |
0.13 |
| chr2_109281814_109282372 | 0.43 |
Kif18a |
kinesin family member 18A |
1224 |
0.4 |
| chr3_127931525_127931952 | 0.43 |
9830132P13Rik |
RIKEN cDNA 9830132P13 gene |
15566 |
0.14 |
| chr3_4796861_4798079 | 0.42 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
88 |
0.98 |
| chr11_95012524_95013312 | 0.42 |
Samd14 |
sterile alpha motif domain containing 14 |
2637 |
0.16 |
| chr15_80081033_80081744 | 0.41 |
Rpl3 |
ribosomal protein L3 |
907 |
0.27 |
| chr11_103102696_103105788 | 0.39 |
Acbd4 |
acyl-Coenzyme A binding domain containing 4 |
463 |
0.7 |
| chr2_155990244_155990644 | 0.38 |
Cep250 |
centrosomal protein 250 |
4529 |
0.12 |
| chr6_87765679_87766248 | 0.35 |
Gm5879 |
predicted gene 5879 |
241 |
0.82 |
| chr2_26576901_26577217 | 0.34 |
Egfl7 |
EGF-like domain 7 |
2955 |
0.12 |
| chr2_30185302_30185471 | 0.34 |
Spout1 |
SPOUT domain containing methyltransferase 1 |
6927 |
0.1 |
| chr13_115529338_115529813 | 0.33 |
Gm6035 |
predicted gene 6035 |
197 |
0.97 |
| chr12_17539903_17540224 | 0.33 |
Odc1 |
ornithine decarboxylase, structural 1 |
4731 |
0.16 |
| chr1_97656733_97656884 | 0.32 |
AC099860.1 |
proline rich protein BstNI subfamily 4 (PRB4), pseudogene |
4023 |
0.23 |
| chr15_45112888_45113052 | 0.32 |
Kcnv1 |
potassium channel, subfamily V, member 1 |
1857 |
0.43 |
| chr4_154024246_154024397 | 0.32 |
Smim1 |
small integral membrane protein 1 |
2 |
0.95 |
| chr19_55125788_55126185 | 0.31 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
1217 |
0.49 |
| chr13_37826269_37827832 | 0.31 |
Rreb1 |
ras responsive element binding protein 1 |
118 |
0.96 |
| chr9_95554372_95554523 | 0.31 |
Gm32281 |
predicted gene, 32281 |
50 |
0.96 |
| chr4_112955735_112956277 | 0.31 |
Gm12816 |
predicted gene 12816 |
280 |
0.92 |
| chr13_98810152_98810868 | 0.30 |
Fcho2 |
FCH domain only 2 |
4643 |
0.14 |
| chr18_21283979_21284139 | 0.30 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
16064 |
0.17 |
| chr9_66182641_66182896 | 0.30 |
Dapk2 |
death-associated protein kinase 2 |
24533 |
0.17 |
| chrX_13202535_13203215 | 0.29 |
Rpl3-ps1 |
ribosomal protein L3, pseudogene 1 |
304 |
0.82 |
| chr5_150642312_150642463 | 0.29 |
N4bp2l2 |
NEDD4 binding protein 2-like 2 |
5132 |
0.13 |
| chr15_76666348_76670076 | 0.29 |
Foxh1 |
forkhead box H1 |
1590 |
0.15 |
| chr12_95693703_95697493 | 0.28 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
241 |
0.92 |
| chr11_69945712_69946013 | 0.28 |
Slc2a4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
2112 |
0.1 |
| chr16_56036340_56036491 | 0.27 |
Gm22422 |
predicted gene, 22422 |
528 |
0.58 |
| chr15_80098310_80099146 | 0.27 |
Syngr1 |
synaptogyrin 1 |
858 |
0.41 |
| chr15_36606762_36607275 | 0.26 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
1552 |
0.29 |
| chr1_131274859_131275440 | 0.26 |
Ikbke |
inhibitor of kappaB kinase epsilon |
1564 |
0.27 |
| chr12_111833317_111833616 | 0.25 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
8701 |
0.11 |
| chr8_94174521_94174806 | 0.25 |
Gm45774 |
predicted gene 45774 |
1251 |
0.25 |
| chrX_140956935_140957373 | 0.25 |
Atg4a |
autophagy related 4A, cysteine peptidase |
204 |
0.73 |
| chr9_103228079_103228881 | 0.24 |
Trf |
transferrin |
38 |
0.97 |
| chr11_84701121_84702313 | 0.24 |
1700109G15Rik |
RIKEN cDNA 1700109G15 gene |
18715 |
0.2 |
| chr4_155847422_155848255 | 0.24 |
Dvl1 |
dishevelled segment polarity protein 1 |
167 |
0.85 |
| chr13_23695605_23696868 | 0.23 |
H1f6 |
H1.6 linker histone, cluster member |
422 |
0.53 |
| chr5_67845808_67845959 | 0.23 |
Gm15477 |
predicted gene 15477 |
1182 |
0.26 |
| chr1_178321962_178322153 | 0.23 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
2114 |
0.17 |
| chr8_13038721_13039033 | 0.22 |
F10 |
coagulation factor X |
1039 |
0.34 |
| chr6_54008580_54009014 | 0.21 |
4921529L05Rik |
RIKEN cDNA 4921529L05 gene |
915 |
0.6 |
| chr6_119009028_119009204 | 0.21 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
70891 |
0.12 |
| chr4_123286782_123287699 | 0.21 |
Gm25788 |
predicted gene, 25788 |
4104 |
0.11 |
| chr17_31057450_31058949 | 0.21 |
Abcg1 |
ATP binding cassette subfamily G member 1 |
425 |
0.72 |
| chr6_8208939_8210019 | 0.20 |
Mios |
meiosis regulator for oocyte development |
257 |
0.91 |
| chr2_181081653_181082036 | 0.20 |
Kcnq2 |
potassium voltage-gated channel, subfamily Q, member 2 |
13577 |
0.13 |
| chr7_120176025_120176210 | 0.20 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
2259 |
0.22 |
| chr4_118080461_118080612 | 0.20 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
48654 |
0.11 |
| chr19_45779130_45779398 | 0.20 |
Oga |
O-GlcNAcase |
4287 |
0.16 |
| chr9_113915715_113915866 | 0.19 |
Ubp1 |
upstream binding protein 1 |
15144 |
0.21 |
| chr1_90216532_90216683 | 0.19 |
Gm38277 |
predicted gene, 38277 |
11919 |
0.16 |
| chr2_57613916_57615034 | 0.19 |
Gm13532 |
predicted gene 13532 |
14753 |
0.2 |
| chr8_109250241_109250821 | 0.19 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
665 |
0.8 |
| chr12_86881882_86884741 | 0.19 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
1487 |
0.4 |
| chr15_61365878_61366029 | 0.19 |
Gm24696 |
predicted gene, 24696 |
7949 |
0.28 |
| chr4_59233833_59234326 | 0.19 |
Ugcg |
UDP-glucose ceramide glucosyltransferase |
21423 |
0.16 |
| chr3_89458801_89459905 | 0.19 |
Pmvk |
phosphomevalonate kinase |
22 |
0.95 |
| chr12_55303747_55303898 | 0.18 |
Prorp |
protein only RNase P catalytic subunit |
536 |
0.54 |
| chr19_9896568_9896747 | 0.18 |
Incenp |
inner centromere protein |
1197 |
0.24 |
| chr2_79268859_79269010 | 0.18 |
Itga4 |
integrin alpha 4 |
12987 |
0.22 |
| chr15_83366703_83367634 | 0.18 |
1700001L05Rik |
RIKEN cDNA 1700001L05 gene |
109 |
0.96 |
| chr3_40745266_40746433 | 0.18 |
Hspa4l |
heat shock protein 4 like |
19 |
0.97 |
| chr1_24349529_24350209 | 0.18 |
Col19a1 |
collagen, type XIX, alpha 1 |
102876 |
0.06 |
| chr2_93456061_93456647 | 0.17 |
Gm10804 |
predicted gene 10804 |
3533 |
0.2 |
| chr5_122291128_122291986 | 0.17 |
Pptc7 |
PTC7 protein phosphatase homolog |
4215 |
0.13 |
| chr6_22878521_22878977 | 0.16 |
Ptprz1 |
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
2972 |
0.31 |
| chr5_36733334_36733545 | 0.16 |
Gm43701 |
predicted gene 43701 |
15179 |
0.12 |
| chr11_85132321_85132613 | 0.16 |
Usp32 |
ubiquitin specific peptidase 32 |
7462 |
0.14 |
| chr8_105855169_105856239 | 0.15 |
Thap11 |
THAP domain containing 11 |
572 |
0.5 |
| chr13_64149393_64149567 | 0.15 |
Zfp367 |
zinc finger protein 367 |
734 |
0.52 |
| chr16_52303393_52303591 | 0.15 |
Alcam |
activated leukocyte cell adhesion molecule |
6568 |
0.33 |
| chr15_74516560_74519631 | 0.15 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
1264 |
0.47 |
| chr15_85677376_85679232 | 0.15 |
Lncppara |
long noncoding RNA near Ppara |
24688 |
0.12 |
| chr15_27994630_27995403 | 0.15 |
Trio |
triple functional domain (PTPRF interacting) |
347 |
0.9 |
| chr3_152198770_152199020 | 0.15 |
Dnajb4 |
DnaJ heat shock protein family (Hsp40) member B4 |
5050 |
0.14 |
| chr10_80141919_80142171 | 0.15 |
Atp5d |
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
270 |
0.76 |
| chr17_46817734_46818115 | 0.14 |
Bicral |
BRD4 interacting chromatin remodeling complex associated protein like |
5739 |
0.14 |
| chr8_35596914_35597077 | 0.14 |
Gm16793 |
predicted gene, 16793 |
8023 |
0.19 |
| chr4_148149332_148150056 | 0.14 |
Fbxo6 |
F-box protein 6 |
25 |
0.95 |
| chr9_94538014_94538635 | 0.14 |
Dipk2a |
divergent protein kinase domain 2A |
243 |
0.94 |
| chr7_144132849_144134003 | 0.14 |
Gm44999 |
predicted gene 44999 |
22341 |
0.2 |
| chr4_145245818_145247120 | 0.14 |
Tnfrsf1b |
tumor necrosis factor receptor superfamily, member 1b |
401 |
0.87 |
| chr17_29660688_29661601 | 0.14 |
Cmtr1 |
cap methyltransferase 1 |
337 |
0.79 |
| chr9_21836571_21836722 | 0.14 |
Angptl8 |
angiopoietin-like 8 |
1136 |
0.32 |
| chr9_71163629_71163837 | 0.14 |
Gm32511 |
predicted gene, 32511 |
61 |
0.87 |
| chr6_120914700_120915049 | 0.14 |
Gm44260 |
predicted gene, 44260 |
119 |
0.93 |
| chr10_111249619_111250191 | 0.13 |
Osbpl8 |
oxysterol binding protein-like 8 |
1837 |
0.35 |
| chr1_91252025_91252209 | 0.13 |
Ube2f |
ubiquitin-conjugating enzyme E2F (putative) |
1461 |
0.34 |
| chr17_33715168_33715319 | 0.13 |
Marchf2 |
membrane associated ring-CH-type finger 2 |
1862 |
0.19 |
| chr2_117440042_117440467 | 0.13 |
Gm13982 |
predicted gene 13982 |
22557 |
0.24 |
| chr18_67563907_67564259 | 0.13 |
Gm17669 |
predicted gene, 17669 |
1689 |
0.34 |
| chr11_102894553_102894704 | 0.13 |
Gfap |
glial fibrillary acidic protein |
2503 |
0.15 |
| chr10_127643084_127644231 | 0.12 |
Stat6 |
signal transducer and activator of transcription 6 |
655 |
0.49 |
| chr10_59800628_59800855 | 0.12 |
Gm17059 |
predicted gene 17059 |
487 |
0.74 |
| chr4_117121385_117121536 | 0.12 |
Btbd19 |
BTB (POZ) domain containing 19 |
195 |
0.83 |
| chr14_31020004_31020155 | 0.12 |
Pbrm1 |
polybromo 1 |
871 |
0.28 |
| chr17_57246789_57247636 | 0.12 |
Gpr108 |
G protein-coupled receptor 108 |
451 |
0.69 |
| chr15_82792748_82793304 | 0.12 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
1196 |
0.3 |
| chr2_77515617_77516160 | 0.12 |
Zfp385b |
zinc finger protein 385B |
3647 |
0.32 |
| chr15_102516242_102517900 | 0.12 |
Map3k12 |
mitogen-activated protein kinase kinase kinase 12 |
7 |
0.93 |
| chr8_10928168_10928319 | 0.12 |
3930402G23Rik |
RIKEN cDNA 3930402G23 gene |
201 |
0.87 |
| chr13_9095816_9096692 | 0.12 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
2272 |
0.25 |
| chr12_40842378_40842529 | 0.11 |
Gm19441 |
predicted gene, 19441 |
33084 |
0.14 |
| chr13_36613015_36613742 | 0.11 |
Gm46409 |
predicted gene, 46409 |
406 |
0.82 |
| chr14_118768312_118768652 | 0.11 |
Gm22379 |
predicted gene, 22379 |
6670 |
0.17 |
| chr2_92458690_92458841 | 0.11 |
Slc35c1 |
solute carrier family 35, member C1 |
944 |
0.45 |
| chr5_124094995_124096382 | 0.11 |
Abcb9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
98 |
0.94 |
| chr2_164912255_164912427 | 0.11 |
Zfp335 |
zinc finger protein 335 |
584 |
0.5 |
| chr11_29527773_29527935 | 0.11 |
Mtif2 |
mitochondrial translational initiation factor 2 |
1397 |
0.27 |
| chr4_134982769_134983159 | 0.11 |
Syf2 |
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
51954 |
0.11 |
| chr2_120404274_120404845 | 0.11 |
Ganc |
glucosidase, alpha; neutral C |
209 |
0.72 |
| chr15_79691713_79691926 | 0.11 |
n-R5s40 |
nuclear encoded rRNA 5S 40 |
683 |
0.38 |
| chr2_162931631_162931821 | 0.11 |
Srsf6 |
serine and arginine-rich splicing factor 6 |
65 |
0.96 |
| chr16_20619702_20620440 | 0.11 |
Camk2n2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
1216 |
0.19 |
| chr12_102757998_102758734 | 0.11 |
Gm20604 |
predicted gene 20604 |
306 |
0.39 |
| chr4_46720937_46721311 | 0.11 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
3267 |
0.31 |
| chr13_51649766_51650138 | 0.11 |
Secisbp2 |
SECIS binding protein 2 |
1745 |
0.26 |
| chr11_83227446_83227655 | 0.10 |
Gm11427 |
predicted gene 11427 |
3064 |
0.12 |
| chr2_18056806_18058206 | 0.10 |
Mir7655 |
microRNA 7655 |
340 |
0.69 |
| chrY_90828377_90829257 | 0.10 |
Gm21742 |
predicted gene, 21742 |
8596 |
0.19 |
| chr15_95702325_95702476 | 0.10 |
Gm8843 |
predicted gene 8843 |
12793 |
0.16 |
| chr7_60960862_60961355 | 0.10 |
Gm44643 |
predicted gene 44643 |
277 |
0.94 |
| chr3_30407909_30408060 | 0.10 |
Gm37024 |
predicted gene, 37024 |
80442 |
0.07 |
| chr7_25176596_25177413 | 0.10 |
Pou2f2 |
POU domain, class 2, transcription factor 2 |
2722 |
0.15 |
| chr19_57357866_57358369 | 0.10 |
Fam160b1 |
family with sequence similarity 160, member B1 |
2563 |
0.21 |
| chr4_137927893_137928360 | 0.10 |
Ece1 |
endothelin converting enzyme 1 |
14444 |
0.2 |
| chr8_94874074_94874812 | 0.10 |
Dok4 |
docking protein 4 |
1653 |
0.23 |
| chr1_69759441_69760065 | 0.10 |
Gm37930 |
predicted gene, 37930 |
11115 |
0.24 |
| chr19_46396930_46398133 | 0.10 |
Sufu |
SUFU negative regulator of hedgehog signaling |
505 |
0.64 |
| chr7_61980139_61980379 | 0.10 |
A330076H08Rik |
RIKEN cDNA A330076H08 gene |
2044 |
0.2 |
| chr11_101936838_101937005 | 0.10 |
Rpl27-ps2 |
ribosomal protein L27, pseudogene 2 |
2580 |
0.16 |
| chr1_173366699_173367980 | 0.09 |
Cadm3 |
cell adhesion molecule 3 |
294 |
0.89 |
| chr11_97745312_97745500 | 0.09 |
Mir8102 |
microRNA 8102 |
509 |
0.43 |
| chr3_145923629_145924032 | 0.09 |
Bcl10 |
B cell leukemia/lymphoma 10 |
432 |
0.82 |
| chr15_82243863_82244385 | 0.09 |
Cenpm |
centromere protein M |
208 |
0.85 |
| chr4_133016211_133017810 | 0.09 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
4806 |
0.2 |
| chr10_84917701_84918233 | 0.09 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
311 |
0.93 |
| chr8_94035053_94035555 | 0.09 |
Nudt21 |
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
1702 |
0.22 |
| chr1_64731523_64731946 | 0.09 |
Gm38058 |
predicted gene, 38058 |
4506 |
0.18 |
| chr11_77800451_77802048 | 0.09 |
Myo18a |
myosin XVIIIA |
49 |
0.97 |
| chr4_100123667_100123818 | 0.09 |
Gm12701 |
predicted gene 12701 |
4121 |
0.26 |
| chr18_76469370_76469521 | 0.09 |
Gm50360 |
predicted gene, 50360 |
147621 |
0.04 |
| chr2_25290133_25293095 | 0.09 |
Grin1os |
glutamate receptor, ionotropic, NMDA1 (zeta 1), opposite strand |
394 |
0.57 |
| chr19_41345332_41345831 | 0.09 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
39515 |
0.17 |
| chr11_117115216_117116282 | 0.09 |
Sec14l1 |
SEC14-like lipid binding 1 |
497 |
0.68 |
| chr3_89225214_89225991 | 0.08 |
Thbs3 |
thrombospondin 3 |
425 |
0.52 |
| chr2_122364330_122364498 | 0.08 |
Shf |
Src homology 2 domain containing F |
2414 |
0.2 |
| chr13_117605053_117605376 | 0.08 |
Hcn1 |
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
2734 |
0.39 |
| chr2_73910369_73911256 | 0.08 |
Atp5g3 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
353 |
0.88 |
| chr15_58933816_58934225 | 0.08 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
212 |
0.57 |
| chr8_76898913_76900466 | 0.08 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
83 |
0.77 |
| chr7_19562594_19562787 | 0.08 |
Ppp1r37 |
protein phosphatase 1, regulatory subunit 37 |
386 |
0.71 |
| chr16_36453993_36454231 | 0.08 |
Stfa3 |
stefin A3 |
1280 |
0.27 |
| chr11_84592794_84593319 | 0.08 |
Lhx1os |
LIM homeobox 1, opposite strand |
67372 |
0.1 |
| chr5_38503165_38503316 | 0.08 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
97 |
0.96 |
| chr7_29308550_29309973 | 0.08 |
Dpf1 |
D4, zinc and double PHD fingers family 1 |
252 |
0.86 |
| chr6_83325542_83325912 | 0.08 |
Mthfd2 |
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
181 |
0.62 |
| chr13_81783279_81784146 | 0.07 |
Gm17259 |
predicted gene, 17259 |
37 |
0.6 |
| chr14_12191443_12191651 | 0.07 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
1604 |
0.41 |
| chrX_162017252_162017925 | 0.07 |
Gm26317 |
predicted gene, 26317 |
51869 |
0.15 |
| chr8_25753750_25754361 | 0.07 |
Ddhd2 |
DDHD domain containing 2 |
225 |
0.66 |
| chr16_35810940_35811264 | 0.07 |
Gm26838 |
predicted gene, 26838 |
2156 |
0.22 |
| chr15_76713644_76714446 | 0.07 |
Lrrc14 |
leucine rich repeat containing 14 |
3314 |
0.09 |
| chr14_121359067_121359218 | 0.07 |
Stk24 |
serine/threonine kinase 24 |
726 |
0.7 |
| chr7_130367477_130367756 | 0.07 |
Gm5903 |
predicted gene 5903 |
13254 |
0.22 |
| chr11_101277950_101278906 | 0.07 |
Coa3 |
cytochrome C oxidase assembly factor 3 |
515 |
0.36 |
| chr11_93103184_93103429 | 0.07 |
Car10 |
carbonic anhydrase 10 |
4016 |
0.37 |
| chr19_8710109_8710483 | 0.07 |
Slc3a2 |
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
2689 |
0.07 |
| chr8_117261523_117261917 | 0.07 |
Cmip |
c-Maf inducing protein |
4603 |
0.29 |
| chr5_124048455_124048606 | 0.07 |
Gm43661 |
predicted gene 43661 |
3822 |
0.12 |
| chr10_69351583_69352885 | 0.07 |
Cdk1 |
cyclin-dependent kinase 1 |
669 |
0.68 |
| chr18_60927346_60927498 | 0.07 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
473 |
0.75 |
| chr1_72710929_72711175 | 0.07 |
Rpl37a |
ribosomal protein L37a |
238 |
0.9 |
| chr4_117891166_117892323 | 0.07 |
Dph2 |
DPH2 homolog |
242 |
0.84 |
| chr2_167337310_167337492 | 0.07 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
11782 |
0.18 |
| chr1_81133034_81133185 | 0.07 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
55526 |
0.17 |
| chr10_79681285_79681959 | 0.07 |
Cdc34 |
cell division cycle 34 |
573 |
0.51 |
| chr9_88520550_88520755 | 0.06 |
Snord50a |
small nucleolar RNA, C/D box 50A |
948 |
0.28 |
| chr16_96122312_96122501 | 0.06 |
Hmgn1 |
high mobility group nucleosomal binding domain 1 |
2933 |
0.17 |
| chr18_9449055_9449747 | 0.06 |
Ccny |
cyclin Y |
285 |
0.9 |
| chr16_3907313_3907961 | 0.06 |
1700037C18Rik |
RIKEN cDNA 1700037C18 gene |
14 |
0.91 |
| chr7_46639032_46639734 | 0.06 |
Sergef |
secretion regulating guanine nucleotide exchange factor |
197 |
0.91 |
| chr11_115698285_115698728 | 0.06 |
Grb2 |
growth factor receptor bound protein 2 |
801 |
0.46 |
| chr19_41906848_41907300 | 0.06 |
Gm46644 |
predicted gene, 46644 |
4185 |
0.12 |
| chr1_66386919_66387899 | 0.06 |
Map2 |
microtubule-associated protein 2 |
398 |
0.87 |
| chr17_46282048_46282988 | 0.06 |
Tjap1 |
tight junction associated protein 1 |
138 |
0.92 |
| chr7_107594370_107595799 | 0.06 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
123 |
0.95 |
| chr17_17294036_17294239 | 0.06 |
Gm6712 |
predicted gene 6712 |
306 |
0.9 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.3 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0071351 | cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.0 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |