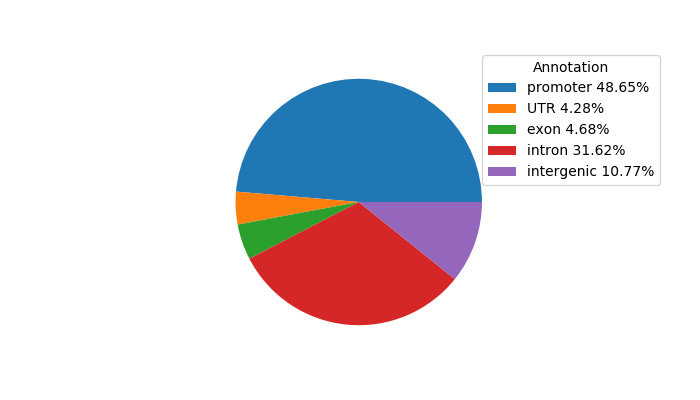
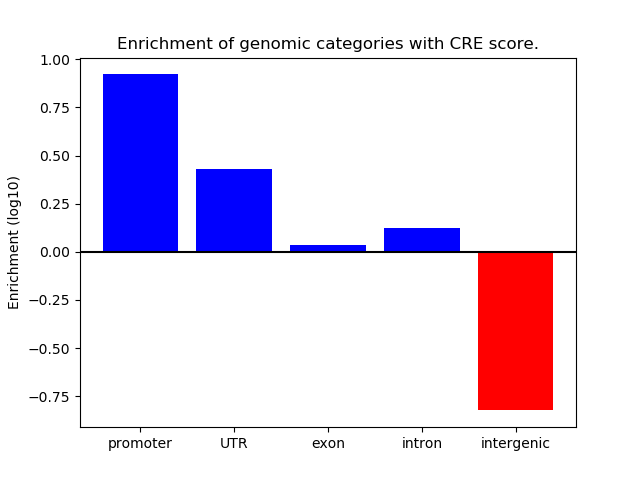
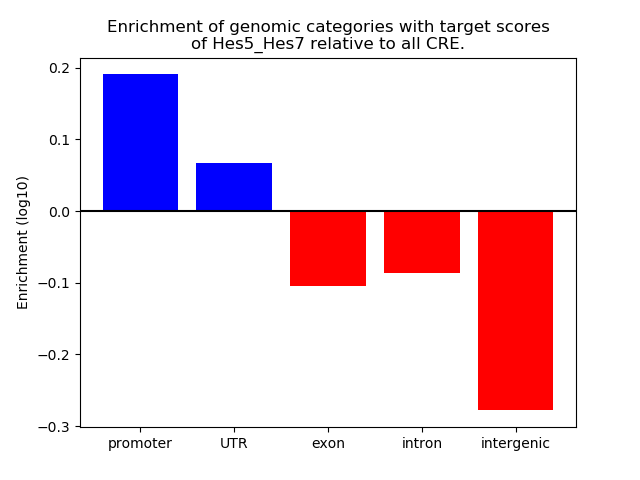
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
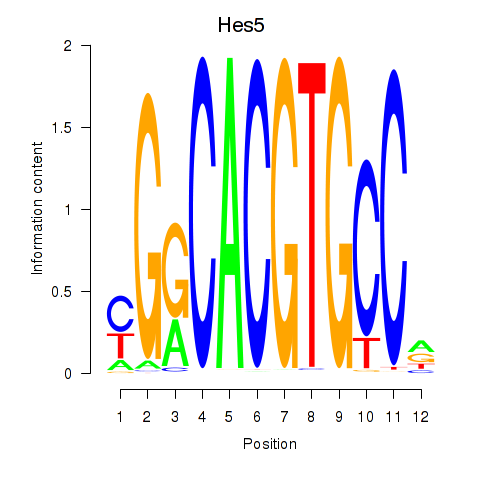
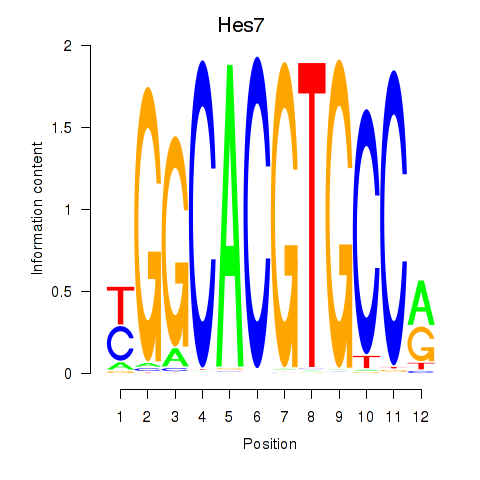
Results for Hes5_Hes7
Z-value: 0.86
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSMUSG00000048001.7 | hes family bHLH transcription factor 5 |
|
Hes7
|
ENSMUSG00000023781.2 | hes family bHLH transcription factor 7 |
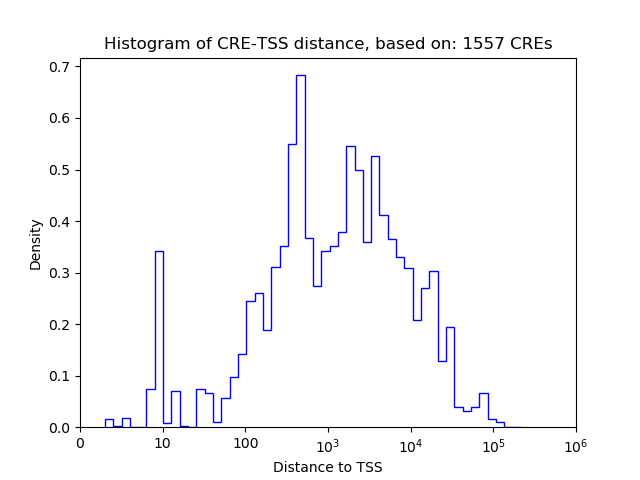
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_154945869_154946537 | Hes5 | 14720 | 0.105177 | -0.47 | 1.4e-04 | Click! |
| chr4_154952657_154953434 | Hes5 | 7878 | 0.115122 | -0.42 | 7.8e-04 | Click! |
| chr4_154946573_154946849 | Hes5 | 14212 | 0.105813 | -0.41 | 1.1e-03 | Click! |
| chr4_154949467_154949911 | Hes5 | 11234 | 0.109628 | -0.36 | 5.2e-03 | Click! |
| chr4_154957968_154959029 | Hes5 | 2425 | 0.170866 | -0.31 | 1.6e-02 | Click! |
| chr11_69116585_69116736 | Hes7 | 3744 | 0.092076 | -0.42 | 9.6e-04 | Click! |
| chr11_69110534_69110843 | Hes7 | 9716 | 0.070699 | -0.41 | 1.0e-03 | Click! |
| chr11_69120197_69120778 | Hes7 | 83 | 0.918272 | -0.41 | 1.1e-03 | Click! |
| chr11_69110991_69111333 | Hes7 | 9242 | 0.071417 | -0.41 | 1.3e-03 | Click! |
| chr11_69120854_69121005 | Hes7 | 525 | 0.529244 | -0.38 | 2.5e-03 | Click! |
Activity of the Hes5_Hes7 motif across conditions
Conditions sorted by the z-value of the Hes5_Hes7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
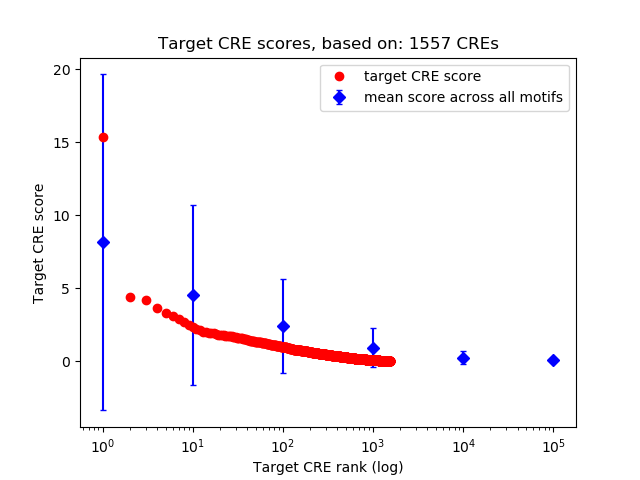
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_12025582_12027083 | 15.35 |
Grb10 |
growth factor receptor bound protein 10 |
8 |
0.98 |
| chr5_103756323_103756745 | 4.41 |
Aff1 |
AF4/FMR2 family, member 1 |
1961 |
0.36 |
| chrX_9256153_9256810 | 4.23 |
Gm14862 |
predicted gene 14862 |
418 |
0.78 |
| chr6_146221639_146221985 | 3.67 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
5731 |
0.27 |
| chr4_129463334_129463725 | 3.29 |
Bsdc1 |
BSD domain containing 1 |
1013 |
0.34 |
| chr17_57234704_57235202 | 3.13 |
C3 |
complement component 3 |
6817 |
0.11 |
| chr13_59766981_59767805 | 2.88 |
Isca1 |
iron-sulfur cluster assembly 1 |
2084 |
0.13 |
| chr17_71267820_71268552 | 2.72 |
Emilin2 |
elastin microfibril interfacer 2 |
411 |
0.81 |
| chr12_33317702_33317935 | 2.48 |
Atxn7l1 |
ataxin 7-like 1 |
2413 |
0.31 |
| chr7_126369150_126370664 | 2.35 |
Lat |
linker for activation of T cells |
337 |
0.73 |
| chr11_31824541_31824785 | 2.21 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
139 |
0.96 |
| chr8_105799077_105799898 | 2.12 |
Ranbp10 |
RAN binding protein 10 |
27718 |
0.08 |
| chr4_59924419_59924679 | 2.03 |
Slc46a2 |
solute carrier family 46, member 2 |
9493 |
0.15 |
| chr6_124683434_124683706 | 2.01 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
17849 |
0.06 |
| chr10_75937026_75937671 | 1.95 |
Chchd10 |
coiled-coil-helix-coiled-coil-helix domain containing 10 |
128 |
0.86 |
| chr7_80200552_80200846 | 1.93 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
2163 |
0.17 |
| chrX_8271051_8272966 | 1.92 |
Slc38a5 |
solute carrier family 38, member 5 |
366 |
0.82 |
| chr10_80604324_80605241 | 1.84 |
Gm49322 |
predicted gene, 49322 |
1239 |
0.18 |
| chr11_82845042_82846306 | 1.83 |
Rffl |
ring finger and FYVE like domain containing protein |
418 |
0.75 |
| chr4_148602903_148603733 | 1.82 |
Masp2 |
mannan-binding lectin serine peptidase 2 |
743 |
0.48 |
| chr6_41704510_41704661 | 1.82 |
Kel |
Kell blood group |
246 |
0.88 |
| chr11_114890015_114890857 | 1.82 |
Cd300a |
CD300A molecule |
142 |
0.93 |
| chr11_58641831_58642253 | 1.74 |
Trim58 |
tripartite motif-containing 58 |
1577 |
0.18 |
| chr19_42583444_42583682 | 1.74 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
2402 |
0.27 |
| chr5_92386923_92387342 | 1.73 |
Art3 |
ADP-ribosyltransferase 3 |
245 |
0.87 |
| chr2_74655002_74656520 | 1.73 |
Evx2 |
even-skipped homeobox 2 |
3658 |
0.08 |
| chr16_17802223_17803941 | 1.73 |
Scarf2 |
scavenger receptor class F, member 2 |
320 |
0.79 |
| chr7_88482138_88482595 | 1.66 |
Gm15661 |
predicted gene 15661 |
33136 |
0.16 |
| chr11_103360110_103361992 | 1.65 |
Arhgap27 |
Rho GTPase activating protein 27 |
151 |
0.92 |
| chr8_105822112_105822566 | 1.64 |
Ranbp10 |
RAN binding protein 10 |
4866 |
0.1 |
| chr6_64728581_64730207 | 1.61 |
Atoh1 |
atonal bHLH transcription factor 1 |
269 |
0.95 |
| chr13_55020667_55020922 | 1.60 |
Hk3 |
hexokinase 3 |
110 |
0.96 |
| chr2_163203169_163205126 | 1.59 |
Tox2 |
TOX high mobility group box family member 2 |
406 |
0.85 |
| chr8_120368836_120369597 | 1.57 |
Gm22715 |
predicted gene, 22715 |
74333 |
0.08 |
| chr1_134418556_134420350 | 1.57 |
Adipor1 |
adiponectin receptor 1 |
3889 |
0.13 |
| chr13_58522751_58523890 | 1.55 |
Gm3131 |
predicted gene 3131 |
22708 |
0.16 |
| chr13_47111734_47112164 | 1.53 |
1700026N04Rik |
RIKEN cDNA 1700026N04 gene |
5530 |
0.13 |
| chr1_165818186_165818830 | 1.51 |
Gm23402 |
predicted gene, 23402 |
8766 |
0.1 |
| chr11_95839544_95839887 | 1.51 |
Abi3 |
ABI gene family, member 3 |
2383 |
0.15 |
| chr16_8673428_8673579 | 1.45 |
Carhsp1 |
calcium regulated heat stable protein 1 |
1348 |
0.3 |
| chr5_73629767_73629939 | 1.44 |
Lrrc66 |
leucine rich repeat containing 66 |
459 |
0.78 |
| chr12_100729481_100729632 | 1.43 |
Rps6ka5 |
ribosomal protein S6 kinase, polypeptide 5 |
2573 |
0.23 |
| chr2_181419557_181419895 | 1.42 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
4662 |
0.11 |
| chr3_14878521_14878851 | 1.39 |
Car2 |
carbonic anhydrase 2 |
7587 |
0.19 |
| chr3_89149727_89149878 | 1.39 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
1412 |
0.18 |
| chr11_11685172_11685688 | 1.38 |
Gm11999 |
predicted gene 11999 |
30 |
0.88 |
| chr10_80314572_80317647 | 1.38 |
2310011J03Rik |
RIKEN cDNA 2310011J03 gene |
4428 |
0.07 |
| chr13_75709278_75709841 | 1.37 |
Ell2 |
elongation factor RNA polymerase II 2 |
1848 |
0.23 |
| chr12_112860205_112860664 | 1.36 |
Gpr132 |
G protein-coupled receptor 132 |
518 |
0.63 |
| chr19_32180962_32181132 | 1.35 |
Sgms1 |
sphingomyelin synthase 1 |
15381 |
0.21 |
| chr14_89898179_89899233 | 1.34 |
Gm10110 |
predicted gene 10110 |
240 |
0.96 |
| chr11_117798237_117798922 | 1.32 |
6030468B19Rik |
RIKEN cDNA 6030468B19 gene |
919 |
0.33 |
| chr12_111814961_111815406 | 1.32 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
942 |
0.36 |
| chr6_127151640_127154010 | 1.30 |
Ccnd2 |
cyclin D2 |
632 |
0.57 |
| chr16_30063273_30064853 | 1.30 |
Hes1 |
hes family bHLH transcription factor 1 |
321 |
0.86 |
| chr15_97786454_97786859 | 1.29 |
Slc48a1 |
solute carrier family 48 (heme transporter), member 1 |
2167 |
0.21 |
| chr14_52179104_52179255 | 1.29 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
16049 |
0.09 |
| chr4_149696192_149696820 | 1.27 |
Pik3cd |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
2046 |
0.23 |
| chr11_83281871_83282887 | 1.25 |
Slfn14 |
schlafen 14 |
4347 |
0.1 |
| chr6_142758576_142758727 | 1.23 |
Cmas |
cytidine monophospho-N-acetylneuraminic acid synthetase |
1876 |
0.35 |
| chr7_79272236_79272387 | 1.23 |
Gm31510 |
predicted gene, 31510 |
500 |
0.59 |
| chr7_97735948_97736244 | 1.22 |
Aqp11 |
aquaporin 11 |
1842 |
0.29 |
| chr2_36088291_36088442 | 1.22 |
Lhx6 |
LIM homeobox protein 6 |
5907 |
0.14 |
| chr4_119183706_119183857 | 1.22 |
Ermap |
erythroblast membrane-associated protein |
4966 |
0.1 |
| chr14_75179791_75179942 | 1.22 |
Lcp1 |
lymphocyte cytosolic protein 1 |
3658 |
0.19 |
| chr8_111929938_111930128 | 1.22 |
Tmem231 |
transmembrane protein 231 |
1873 |
0.25 |
| chr10_68132791_68133177 | 1.21 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
3642 |
0.31 |
| chr17_84185673_84188034 | 1.21 |
Zfp36l2 |
zinc finger protein 36, C3H type-like 2 |
1094 |
0.34 |
| chr4_132420507_132420658 | 1.19 |
Phactr4 |
phosphatase and actin regulator 4 |
1859 |
0.19 |
| chr19_46304366_46306224 | 1.18 |
4833438C02Rik |
RIKEN cDNA 4833438C02 gene |
310 |
0.52 |
| chr5_65864197_65865220 | 1.18 |
Rhoh |
ras homolog family member H |
1090 |
0.34 |
| chr11_95341129_95341703 | 1.17 |
Fam117a |
family with sequence similarity 117, member A |
1454 |
0.28 |
| chr5_123094365_123095026 | 1.14 |
Tmem120b |
transmembrane protein 120B |
5206 |
0.1 |
| chr2_93447410_93447951 | 1.13 |
Cd82 |
CD82 antigen |
4999 |
0.18 |
| chr4_141746794_141747616 | 1.13 |
Agmat |
agmatine ureohydrolase (agmatinase) |
533 |
0.68 |
| chr13_76055370_76056809 | 1.13 |
Gpr150 |
G protein-coupled receptor 150 |
907 |
0.5 |
| chr11_98338372_98339597 | 1.12 |
Neurod2 |
neurogenic differentiation 2 |
9336 |
0.09 |
| chr8_95020660_95021163 | 1.11 |
Adgrg3 |
adhesion G protein-coupled receptor G3 |
117 |
0.94 |
| chr5_31094185_31095713 | 1.11 |
Gm9924 |
predicted gene 9924 |
35 |
0.63 |
| chr13_25097547_25097765 | 1.11 |
Dcdc2a |
doublecortin domain containing 2a |
1309 |
0.46 |
| chr15_74933225_74933498 | 1.10 |
Gm39556 |
predicted gene, 39556 |
3924 |
0.11 |
| chr4_119018136_119018630 | 1.10 |
Gm12862 |
predicted gene 12862 |
17898 |
0.09 |
| chr11_51264400_51264551 | 1.09 |
Clk4 |
CDC like kinase 4 |
1289 |
0.3 |
| chr19_45230983_45235468 | 1.08 |
Lbx1 |
ladybird homeobox 1 |
2587 |
0.27 |
| chr5_118482858_118484042 | 1.08 |
Gm15754 |
predicted gene 15754 |
3517 |
0.25 |
| chr4_137752041_137752211 | 1.08 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
4979 |
0.21 |
| chr10_84550138_84550342 | 1.07 |
Platr7 |
pluripotency associated transcript 7 |
3098 |
0.16 |
| chr14_69283473_69284973 | 1.06 |
Gm31748 |
predicted gene, 31748 |
127 |
0.72 |
| chr11_89298118_89300658 | 1.05 |
Nog |
noggin |
2944 |
0.27 |
| chr4_119051580_119051741 | 1.05 |
Gm12866 |
predicted gene 12866 |
17451 |
0.1 |
| chr2_130634489_130636508 | 1.04 |
Lzts3 |
leucine zipper, putative tumor suppressor family member 3 |
2597 |
0.15 |
| chr18_84885276_84885548 | 1.04 |
Cyb5a |
cytochrome b5 type A (microsomal) |
7811 |
0.17 |
| chr15_99664075_99665492 | 1.02 |
Asic1 |
acid-sensing (proton-gated) ion channel 1 |
5585 |
0.1 |
| chr4_140684433_140684890 | 1.02 |
Gm13025 |
predicted gene 13025 |
4908 |
0.14 |
| chr4_123283015_123283683 | 1.01 |
Pabpc4 |
poly(A) binding protein, cytoplasmic 4 |
254 |
0.84 |
| chr8_94990008_94990523 | 1.00 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
4009 |
0.14 |
| chr4_138220887_138221064 | 0.99 |
Hp1bp3 |
heterochromatin protein 1, binding protein 3 |
3681 |
0.14 |
| chr10_94919501_94920004 | 0.99 |
Plxnc1 |
plexin C1 |
2905 |
0.28 |
| chr2_103960605_103960942 | 0.99 |
Lmo2 |
LIM domain only 2 |
2778 |
0.2 |
| chr13_101612295_101612446 | 0.98 |
Gm29341 |
predicted gene 29341 |
6137 |
0.22 |
| chr2_122035758_122036168 | 0.98 |
Eif3j1 |
eukaryotic translation initiation factor 3, subunit J1 |
7311 |
0.16 |
| chr2_180892979_180894605 | 0.97 |
Mir124a-3 |
microRNA 124a-3 |
248 |
0.53 |
| chr15_3271737_3272448 | 0.97 |
Selenop |
selenoprotein P |
7 |
0.98 |
| chr2_34155129_34155306 | 0.96 |
Gm38389 |
predicted gene, 38389 |
43773 |
0.15 |
| chr19_32222165_32222441 | 0.96 |
Sgms1 |
sphingomyelin synthase 1 |
11290 |
0.22 |
| chr3_89391665_89393338 | 0.95 |
Zbtb7b |
zinc finger and BTB domain containing 7B |
432 |
0.43 |
| chr13_119790868_119791564 | 0.95 |
Zfp131 |
zinc finger protein 131 |
327 |
0.81 |
| chr7_99592940_99593230 | 0.95 |
Arrb1 |
arrestin, beta 1 |
1508 |
0.26 |
| chr14_52140471_52142121 | 0.95 |
Rpgrip1 |
retinitis pigmentosa GTPase regulator interacting protein 1 |
4068 |
0.13 |
| chr7_127217459_127218630 | 0.94 |
Septin1 |
septin 1 |
187 |
0.85 |
| chr5_139196714_139196865 | 0.93 |
Sun1 |
Sad1 and UNC84 domain containing 1 |
3848 |
0.17 |
| chr11_32300067_32300964 | 0.93 |
Hbq1a |
hemoglobin, theta 1A |
446 |
0.71 |
| chr16_36278194_36278405 | 0.92 |
Stfa1 |
stefin A1 |
1152 |
0.24 |
| chr18_20642317_20642474 | 0.91 |
Gm16090 |
predicted gene 16090 |
22865 |
0.15 |
| chr6_120179299_120179618 | 0.91 |
Ninj2 |
ninjurin 2 |
14365 |
0.19 |
| chr9_22054329_22055024 | 0.91 |
Elavl3 |
ELAV like RNA binding protein 3 |
2653 |
0.12 |
| chr18_11049995_11051717 | 0.91 |
Gata6os |
GATA binding protein 6, opposite strand |
631 |
0.64 |
| chr13_35740614_35741201 | 0.90 |
Cdyl |
chromodomain protein, Y chromosome-like |
406 |
0.85 |
| chr6_146579499_146579843 | 0.88 |
Fgfr1op2 |
FGFR1 oncogene partner 2 |
1729 |
0.26 |
| chr18_20944519_20944761 | 0.86 |
Rnf125 |
ring finger protein 125 |
15 |
0.98 |
| chr14_69501975_69503232 | 0.85 |
Gm37847 |
predicted gene, 37847 |
3 |
0.73 |
| chr3_135843805_135844446 | 0.84 |
4933401H06Rik |
RIKEN cDNA 4933401H06 gene |
3856 |
0.17 |
| chr1_181638473_181638624 | 0.84 |
Dnah14 |
dynein, axonemal, heavy chain 14 |
14831 |
0.21 |
| chrX_7968242_7968552 | 0.83 |
Gata1 |
GATA binding protein 1 |
487 |
0.61 |
| chr4_11146992_11147797 | 0.83 |
Gm11830 |
predicted gene 11830 |
2684 |
0.17 |
| chr4_63561098_63561515 | 0.83 |
Tmem268 |
transmembrane protein 268 |
946 |
0.42 |
| chr2_121235231_121235799 | 0.83 |
Trp53bp1 |
transformation related protein 53 binding protein 1 |
174 |
0.93 |
| chr13_104140168_104140319 | 0.83 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
30414 |
0.12 |
| chr15_83346375_83346526 | 0.83 |
Arfgap3 |
ADP-ribosylation factor GTPase activating protein 3 |
3724 |
0.19 |
| chr17_36868940_36869130 | 0.82 |
Trim10 |
tripartite motif-containing 10 |
539 |
0.54 |
| chr18_68159709_68160032 | 0.82 |
Gm18149 |
predicted gene, 18149 |
9097 |
0.21 |
| chr7_28597227_28598131 | 0.81 |
Pak4 |
p21 (RAC1) activated kinase 4 |
407 |
0.46 |
| chr19_3334357_3334684 | 0.81 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
795 |
0.52 |
| chr17_84096219_84097195 | 0.80 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
18047 |
0.14 |
| chr11_50222884_50223393 | 0.80 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2197 |
0.17 |
| chr1_75217955_75219308 | 0.80 |
Tuba4a |
tubulin, alpha 4A |
7 |
0.92 |
| chr12_69372255_69373396 | 0.80 |
Gm9887 |
predicted gene 9887 |
357 |
0.62 |
| chr1_180812210_180812890 | 0.79 |
H3f3a |
H3.3 histone A |
984 |
0.32 |
| chr8_120537063_120538443 | 0.79 |
1700016A09Rik |
RIKEN cDNA 1700016A09 gene |
27 |
0.8 |
| chr5_132542051_132543042 | 0.79 |
Auts2 |
autism susceptibility candidate 2 |
111 |
0.94 |
| chr1_183439062_183439515 | 0.78 |
Gm37214 |
predicted gene, 37214 |
18633 |
0.13 |
| chr5_144255271_144256520 | 0.78 |
2900089D17Rik |
RIKEN cDNA 2900089D17 gene |
291 |
0.71 |
| chr4_98923707_98924979 | 0.77 |
Usp1 |
ubiquitin specific peptidase 1 |
379 |
0.86 |
| chr2_132102631_132103055 | 0.77 |
Gm14052 |
predicted gene 14052 |
2974 |
0.23 |
| chr4_148074823_148075738 | 0.77 |
Gm13201 |
predicted gene 13201 |
140 |
0.91 |
| chr11_19964534_19964685 | 0.77 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
10039 |
0.27 |
| chr4_115088735_115089677 | 0.77 |
Pdzk1ip1 |
PDZK1 interacting protein 1 |
163 |
0.94 |
| chr11_110094976_110095336 | 0.77 |
Abca8a |
ATP-binding cassette, sub-family A (ABC1), member 8a |
781 |
0.73 |
| chr2_103899895_103900335 | 0.76 |
Gm13876 |
predicted gene 13876 |
11791 |
0.11 |
| chr14_69280362_69280833 | 0.76 |
Gm20236 |
predicted gene, 20236 |
1543 |
0.21 |
| chr14_69498607_69499086 | 0.76 |
Gm37094 |
predicted gene, 37094 |
1544 |
0.22 |
| chr10_125321650_125321801 | 0.76 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
6810 |
0.24 |
| chr2_132695147_132695871 | 0.76 |
Shld1 |
shieldin complex subunit 1 |
2610 |
0.16 |
| chr11_98746874_98747641 | 0.76 |
Thra |
thyroid hormone receptor alpha |
3024 |
0.13 |
| chr2_3771077_3771340 | 0.75 |
Fam107b |
family with sequence similarity 107, member B |
500 |
0.8 |
| chr4_153957304_153958293 | 0.75 |
A430005L14Rik |
RIKEN cDNA A430005L14 gene |
469 |
0.71 |
| chr1_39476542_39476838 | 0.74 |
Tbc1d8 |
TBC1 domain family, member 8 |
1971 |
0.28 |
| chr10_36974081_36974232 | 0.74 |
Hdac2 |
histone deacetylase 2 |
388 |
0.92 |
| chr11_79992462_79992880 | 0.73 |
Suz12 |
SUZ12 polycomb repressive complex 2 subunit |
435 |
0.82 |
| chr9_36725804_36726577 | 0.73 |
Chek1 |
checkpoint kinase 1 |
184 |
0.91 |
| chr1_58393178_58393943 | 0.73 |
Bzw1 |
basic leucine zipper and W2 domains 1 |
90 |
0.95 |
| chr18_75502739_75503903 | 0.73 |
Gm10532 |
predicted gene 10532 |
11324 |
0.26 |
| chr2_25577192_25580600 | 0.73 |
Ajm1 |
apical junction component 1 |
1001 |
0.25 |
| chrX_103986162_103986488 | 0.72 |
Rlim |
ring finger protein, LIM domain interacting |
5041 |
0.17 |
| chr1_167269734_167270158 | 0.72 |
Uck2 |
uridine-cytidine kinase 2 |
14655 |
0.11 |
| chr17_28824668_28825067 | 0.71 |
Brpf3 |
bromodomain and PHD finger containing, 3 |
841 |
0.42 |
| chr2_30396345_30396496 | 0.71 |
Dolpp1 |
dolichyl pyrophosphate phosphatase 1 |
822 |
0.43 |
| chr10_19578677_19578828 | 0.71 |
Ifngr1 |
interferon gamma receptor 1 |
13197 |
0.18 |
| chr2_26485135_26488628 | 0.71 |
Notch1 |
notch 1 |
16383 |
0.09 |
| chr15_73183514_73184891 | 0.70 |
Ago2 |
argonaute RISC catalytic subunit 2 |
733 |
0.68 |
| chr1_58957001_58957419 | 0.70 |
Trak2 |
trafficking protein, kinesin binding 2 |
10873 |
0.15 |
| chr3_95141194_95141345 | 0.70 |
Tnfaip8l2 |
tumor necrosis factor, alpha-induced protein 8-like 2 |
1091 |
0.22 |
| chr16_91468070_91468519 | 0.70 |
Gm49626 |
predicted gene, 49626 |
3169 |
0.12 |
| chr1_86358619_86359308 | 0.69 |
Ncl |
nucleolin |
6 |
0.89 |
| chr7_46396141_46398423 | 0.69 |
Kcnc1 |
potassium voltage gated channel, Shaw-related subfamily, member 1 |
366 |
0.84 |
| chr7_44853717_44855163 | 0.69 |
Akt1s1 |
AKT1 substrate 1 (proline-rich) |
1869 |
0.12 |
| chr11_53762840_53764187 | 0.69 |
Mir7671 |
microRNA 7671 |
141 |
0.92 |
| chr5_99339481_99340596 | 0.68 |
Gm35394 |
predicted gene, 35394 |
65943 |
0.12 |
| chr17_46162527_46163464 | 0.68 |
Gtpbp2 |
GTP binding protein 2 |
38 |
0.95 |
| chr17_45594287_45595840 | 0.68 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
439 |
0.67 |
| chr17_74290903_74291403 | 0.68 |
Memo1 |
mediator of cell motility 1 |
2505 |
0.2 |
| chr18_37999322_37999473 | 0.68 |
Arap3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
428 |
0.71 |
| chr18_21152486_21153141 | 0.68 |
Gm6378 |
predicted pseudogene 6378 |
75704 |
0.09 |
| chr4_109467800_109467951 | 0.68 |
Rnf11 |
ring finger protein 11 |
8800 |
0.18 |
| chr1_91546109_91547169 | 0.67 |
Asb1 |
ankyrin repeat and SOCS box-containing 1 |
5559 |
0.17 |
| chr10_75935429_75936681 | 0.67 |
Mmp11 |
matrix metallopeptidase 11 |
441 |
0.42 |
| chr17_47593570_47594831 | 0.67 |
Ccnd3 |
cyclin D3 |
113 |
0.94 |
| chr16_20097588_20098652 | 0.67 |
Klhl24 |
kelch-like 24 |
528 |
0.79 |
| chr19_7056335_7057882 | 0.66 |
Macrod1 |
mono-ADP ribosylhydrolase 1 |
298 |
0.81 |
| chr7_144489028_144489179 | 0.66 |
Ppfia1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
234 |
0.92 |
| chrX_106135756_106136481 | 0.66 |
Tlr13 |
toll-like receptor 13 |
7086 |
0.16 |
| chr11_115491138_115491838 | 0.65 |
Nt5c |
5',3'-nucleotidase, cytosolic |
117 |
0.9 |
| chr5_139791597_139792651 | 0.65 |
Mafk |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
590 |
0.63 |
| chr11_60932121_60933057 | 0.65 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
523 |
0.52 |
| chr3_86224267_86224919 | 0.65 |
Lrba |
LPS-responsive beige-like anchor |
87 |
0.96 |
| chr2_166553882_166554138 | 0.64 |
Prex1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
18967 |
0.19 |
| chr2_131178768_131180148 | 0.64 |
Cenpb |
centromere protein B |
609 |
0.54 |
| chr11_60931035_60931321 | 0.64 |
Map2k3os |
mitogen-activated protein kinase kinase 3, opposite strand |
689 |
0.44 |
| chr11_7197824_7198759 | 0.64 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
509 |
0.77 |
| chr2_127335796_127337945 | 0.64 |
Dusp2 |
dual specificity phosphatase 2 |
260 |
0.86 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.9 | 2.6 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.6 | 1.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.5 | 1.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 1.3 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 | 0.7 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 | 1.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.2 | 1.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 1.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.2 | 0.6 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 0.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 0.7 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.2 | 1.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 0.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.5 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.5 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 0.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 1.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.4 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.5 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.3 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.8 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.3 | GO:0070662 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.3 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.3 | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death(GO:1901298) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.1 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.1 | 0.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.2 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 2.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.4 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.1 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.4 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.4 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.1 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.1 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.6 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 1.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.6 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.0 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.0 | 0.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.0 | 0.1 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.0 | 0.3 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.0 | 0.2 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 1.3 | GO:0043487 | regulation of RNA stability(GO:0043487) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.2 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.0 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.0 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.7 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.0 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.4 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.0 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.0 | GO:1902750 | negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.0 | GO:0034447 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.1 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 1.0 | GO:1901800 | positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.0 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:2000674 | regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.0 | 0.0 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.0 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.2 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.0 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.0 | GO:0072757 | cellular response to camptothecin(GO:0072757) response to camptothecin(GO:1901563) |
| 0.0 | 0.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 | 0.0 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.0 | 0.0 | GO:0051305 | meiotic chromosome movement towards spindle pole(GO:0016344) chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.0 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.2 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 0.4 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 2.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 1.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 16.9 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.4 | 1.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 1.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.4 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.2 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 1.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.3 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.1 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.4 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.6 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0070635 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 2.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 1.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 14.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 2.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |