Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
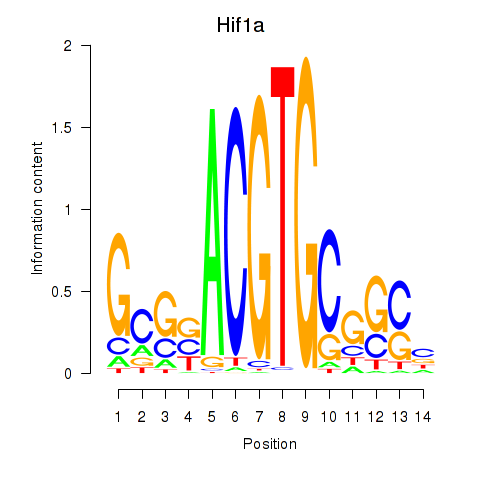
Results for Hif1a
Z-value: 0.54
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.7 | hypoxia inducible factor 1, alpha subunit |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_73901338_73901489 | Hif1a | 38 | 0.973379 | -0.25 | 5.2e-02 | Click! |
| chr12_73916045_73916196 | Hif1a | 8216 | 0.158013 | 0.24 | 6.9e-02 | Click! |
| chr12_73912214_73912365 | Hif1a | 4385 | 0.185784 | 0.15 | 2.5e-01 | Click! |
| chr12_73913244_73913533 | Hif1a | 5484 | 0.173465 | 0.11 | 4.1e-01 | Click! |
| chr12_73902065_73902216 | Hif1a | 765 | 0.619039 | 0.11 | 4.1e-01 | Click! |
Activity of the Hif1a motif across conditions
Conditions sorted by the z-value of the Hif1a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
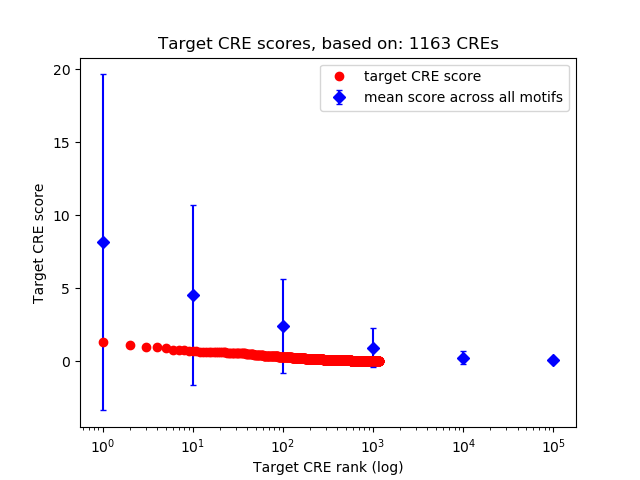
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_118568846_118570341 | 1.29 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
317 |
0.91 |
| chr13_54765549_54766801 | 1.09 |
Sncb |
synuclein, beta |
78 |
0.95 |
| chr7_44336079_44337576 | 0.96 |
Shank1 |
SH3 and multiple ankyrin repeat domains 1 |
811 |
0.34 |
| chr2_152081612_152083149 | 0.95 |
Scrt2 |
scratch family zinc finger 2 |
851 |
0.52 |
| chrX_110814269_110815716 | 0.88 |
Pou3f4 |
POU domain, class 3, transcription factor 4 |
712 |
0.71 |
| chr7_121391567_121393161 | 0.78 |
Hs3st2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
74 |
0.93 |
| chr1_42697532_42698715 | 0.77 |
Pou3f3 |
POU domain, class 3, transcription factor 3 |
2355 |
0.2 |
| chr4_138394975_138396803 | 0.75 |
Fam43b |
family with sequence similarity 43, member B |
639 |
0.41 |
| chr7_87584098_87584781 | 0.71 |
Grm5 |
glutamate receptor, metabotropic 5 |
41 |
0.99 |
| chr11_3331744_3332343 | 0.70 |
Pik3ip1 |
phosphoinositide-3-kinase interacting protein 1 |
29 |
0.93 |
| chr14_3571642_3572719 | 0.67 |
Gm3005 |
predicted gene 3005 |
157 |
0.94 |
| chr19_44674504_44675304 | 0.66 |
Gm26644 |
predicted gene, 26644 |
20371 |
0.15 |
| chr2_22895605_22896237 | 0.66 |
Pdss1 |
prenyl (solanesyl) diphosphate synthase, subunit 1 |
127 |
0.58 |
| chr13_116302578_116304501 | 0.65 |
Isl1 |
ISL1 transcription factor, LIM/homeodomain |
188 |
0.96 |
| chr14_3809696_3810741 | 0.65 |
Gm3002 |
predicted gene 3002 |
144 |
0.94 |
| chr14_5812168_5813063 | 0.64 |
Gm3383 |
predicted gene 3383 |
4633 |
0.14 |
| chr14_4018473_4019517 | 0.63 |
Gm5796 |
predicted gene 5796 |
4946 |
0.14 |
| chr1_91344585_91345841 | 0.63 |
Klhl30 |
kelch-like 30 |
5803 |
0.11 |
| chr14_19584548_19585598 | 0.63 |
Gm2237 |
predicted gene 2237 |
62 |
0.95 |
| chr8_119394920_119395525 | 0.62 |
Mlycd |
malonyl-CoA decarboxylase |
324 |
0.87 |
| chr19_5695606_5696740 | 0.62 |
Map3k11 |
mitogen-activated protein kinase kinase kinase 11 |
3278 |
0.08 |
| chr14_4726384_4727541 | 0.62 |
Gm3252 |
predicted gene 3252 |
187 |
0.91 |
| chr14_6756399_6757462 | 0.61 |
Gm3636 |
predicted gene 3636 |
14598 |
0.14 |
| chr14_4498814_4499998 | 0.60 |
Gm3173 |
predicted gene 3173 |
15352 |
0.11 |
| chr14_6286765_6287923 | 0.60 |
Gm3411 |
predicted gene 3411 |
94 |
0.95 |
| chr7_49911362_49912424 | 0.60 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
193 |
0.96 |
| chr14_3412245_3413254 | 0.58 |
Gm10409 |
predicted gene 10409 |
135 |
0.93 |
| chr15_80287040_80287449 | 0.57 |
Cacna1i |
calcium channel, voltage-dependent, alpha 1I subunit |
6 |
0.97 |
| chr14_5459633_5460571 | 0.57 |
Gm3194 |
predicted gene 3194 |
4635 |
0.14 |
| chr9_87015230_87016430 | 0.57 |
Ripply2 |
ripply transcriptional repressor 2 |
194 |
0.94 |
| chr14_7026733_7027810 | 0.56 |
Gm10406 |
predicted gene 10406 |
178 |
0.93 |
| chr4_22486897_22487389 | 0.56 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
1223 |
0.42 |
| chr9_45430098_45431532 | 0.56 |
4833428L15Rik |
RIKEN cDNA 4833428L15 gene |
417 |
0.52 |
| chr10_81272581_81272732 | 0.56 |
Apba3 |
amyloid beta (A4) precursor protein-binding, family A, member 3 |
38 |
0.9 |
| chr14_6426291_6427481 | 0.55 |
Lamtor3-ps |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3, pseudogene |
14404 |
0.14 |
| chr2_128758874_128759147 | 0.54 |
Gm14011 |
predicted gene 14011 |
5305 |
0.16 |
| chr14_7173946_7175014 | 0.54 |
Gm3512 |
predicted gene 3512 |
53 |
0.97 |
| chr14_3332235_3333462 | 0.54 |
Gm2956 |
predicted gene 2956 |
221 |
0.91 |
| chr14_7487792_7489005 | 0.52 |
Gm3752 |
predicted gene 3752 |
4636 |
0.16 |
| chr14_4182195_4183376 | 0.52 |
Gm2974 |
predicted gene 2974 |
209 |
0.91 |
| chrX_136666183_136667703 | 0.52 |
Tceal3 |
transcription elongation factor A (SII)-like 3 |
332 |
0.84 |
| chr14_7657902_7658985 | 0.51 |
Gm10128 |
predicted gene 10128 |
14471 |
0.16 |
| chr14_5501278_5502453 | 0.50 |
Gm3488 |
predicted gene, 3488 |
191 |
0.9 |
| chr7_46348707_46349190 | 0.50 |
1700025L06Rik |
RIKEN cDNA 1700025L06 gene |
106 |
0.96 |
| chr4_107823086_107824019 | 0.49 |
Lrp8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
391 |
0.53 |
| chr3_73053153_73054210 | 0.48 |
Slitrk3 |
SLIT and NTRK-like family, member 3 |
3262 |
0.23 |
| chr2_164967685_164969910 | 0.47 |
Slc12a5 |
solute carrier family 12, member 5 |
516 |
0.7 |
| chr14_5148637_5149909 | 0.46 |
Gm3317 |
predicted gene 3317 |
15262 |
0.11 |
| chr17_51461105_51461697 | 0.45 |
Gm46574 |
predicted gene, 46574 |
23320 |
0.19 |
| chr7_98193340_98194780 | 0.45 |
B3gnt6 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
5421 |
0.16 |
| chr14_7104815_7105770 | 0.45 |
Gm3696 |
predicted gene 3696 |
4671 |
0.16 |
| chr16_8424071_8425359 | 0.45 |
Tmem114 |
transmembrane protein 114 |
421 |
0.83 |
| chr14_6107781_6108962 | 0.45 |
Gm3468 |
predicted gene 3468 |
294 |
0.84 |
| chr7_137318563_137320070 | 0.44 |
Ebf3 |
early B cell factor 3 |
4871 |
0.21 |
| chr14_4334314_4335468 | 0.44 |
2610042L04Rik |
RIKEN cDNA 2610042L04 gene |
128 |
0.95 |
| chr14_3651660_3652927 | 0.42 |
Gm3020 |
predicted gene 3020 |
263 |
0.87 |
| chr7_79504311_79505700 | 0.42 |
Mir9-3 |
microRNA 9-3 |
259 |
0.82 |
| chr1_177446374_177448525 | 0.41 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
1628 |
0.31 |
| chr5_37241461_37244349 | 0.40 |
Crmp1 |
collapsin response mediator protein 1 |
171 |
0.95 |
| chr11_69800783_69801791 | 0.40 |
Fgf11 |
fibroblast growth factor 11 |
429 |
0.56 |
| chr6_48628821_48630012 | 0.39 |
AI854703 |
expressed sequence AI854703 |
1481 |
0.18 |
| chr14_65423052_65425451 | 0.39 |
Pnoc |
prepronociceptin |
909 |
0.6 |
| chr2_156613822_156614400 | 0.38 |
Dlgap4 |
DLG associated protein 4 |
406 |
0.56 |
| chr11_47988311_47989415 | 0.38 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
106 |
0.98 |
| chr16_43979809_43980249 | 0.38 |
Zdhhc23 |
zinc finger, DHHC domain containing 23 |
238 |
0.91 |
| chr13_39653133_39653284 | 0.38 |
Gm32243 |
predicted gene, 32243 |
5961 |
0.22 |
| chr2_156614815_156615561 | 0.38 |
Gm14174 |
predicted gene 14174 |
260 |
0.82 |
| chr11_82388570_82389922 | 0.37 |
Tmem132e |
transmembrane protein 132E |
123 |
0.96 |
| chr2_25318080_25319601 | 0.37 |
Grin1 |
glutamate receptor, ionotropic, NMDA1 (zeta 1) |
255 |
0.77 |
| chr14_4855192_4856263 | 0.37 |
Gm3264 |
predicted gene 3264 |
151 |
0.94 |
| chr7_44985301_44986386 | 0.37 |
Prmt1 |
protein arginine N-methyltransferase 1 |
473 |
0.48 |
| chr5_139546076_139547826 | 0.37 |
Uncx |
UNC homeobox |
3053 |
0.23 |
| chr18_65430358_65430843 | 0.36 |
Malt1 |
MALT1 paracaspase |
338 |
0.82 |
| chr3_144396551_144396705 | 0.36 |
Gm5857 |
predicted gene 5857 |
31044 |
0.17 |
| chr14_32462734_32463939 | 0.36 |
Chat |
choline acetyltransferase |
1294 |
0.28 |
| chr8_41054476_41055299 | 0.36 |
Mtus1 |
mitochondrial tumor suppressor 1 |
93 |
0.95 |
| chr5_122950311_122951097 | 0.35 |
Kdm2b |
lysine (K)-specific demethylase 2B |
1286 |
0.34 |
| chr11_68384940_68385629 | 0.35 |
Ntn1 |
netrin 1 |
1542 |
0.44 |
| chr12_112928791_112929397 | 0.35 |
Jag2 |
jagged 2 |
646 |
0.51 |
| chr11_11113816_11114856 | 0.34 |
Vwc2 |
von Willebrand factor C domain containing 2 |
113 |
0.98 |
| chr2_70561988_70564432 | 0.34 |
Gad1os |
glutamate decarboxylase 1, opposite strand |
147 |
0.61 |
| chr7_25180336_25182324 | 0.34 |
Pou2f2 |
POU domain, class 2, transcription factor 2 |
1604 |
0.22 |
| chr5_120409346_120410480 | 0.34 |
Lhx5 |
LIM homeobox protein 5 |
21786 |
0.12 |
| chr8_29218667_29220085 | 0.34 |
Unc5d |
unc-5 netrin receptor D |
51 |
0.98 |
| chr10_84754760_84756248 | 0.34 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
558 |
0.8 |
| chr5_110652833_110653341 | 0.34 |
Noc4l |
NOC4 like |
172 |
0.69 |
| chr14_6889307_6890296 | 0.33 |
Gm3667 |
predicted gene 3667 |
161 |
0.95 |
| chr1_17097801_17097987 | 0.33 |
Jph1 |
junctophilin 1 |
5 |
0.98 |
| chr9_37433470_37434351 | 0.33 |
Robo3 |
roundabout guidance receptor 3 |
664 |
0.59 |
| chr18_42394146_42396205 | 0.33 |
Pou4f3 |
POU domain, class 4, transcription factor 3 |
100 |
0.97 |
| chr9_90162957_90164072 | 0.33 |
Adamts7 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
288 |
0.88 |
| chr13_95989973_95990124 | 0.32 |
Sv2c |
synaptic vesicle glycoprotein 2c |
178 |
0.96 |
| chr4_82513112_82513640 | 0.32 |
Gm11266 |
predicted gene 11266 |
5360 |
0.22 |
| chr11_119190727_119190895 | 0.32 |
Gm11753 |
predicted gene 11753 |
6295 |
0.13 |
| chr7_99182821_99183252 | 0.32 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
317 |
0.84 |
| chr4_42917314_42917764 | 0.31 |
Phf24 |
PHD finger protein 24 |
205 |
0.91 |
| chr7_78989959_78991095 | 0.31 |
Gm26633 |
predicted gene, 26633 |
62796 |
0.08 |
| chr3_54915029_54916379 | 0.31 |
Sertm1 |
serine rich and transmembrane domain containing 1 |
183 |
0.96 |
| chr9_91363965_91365514 | 0.31 |
Zic1 |
zinc finger protein of the cerebellum 1 |
1029 |
0.35 |
| chr2_6322398_6322669 | 0.31 |
Usp6nl |
USP6 N-terminal like |
134 |
0.86 |
| chr3_139205663_139206386 | 0.31 |
Stpg2 |
sperm tail PG rich repeat containing 2 |
131 |
0.98 |
| chr13_52529695_52530820 | 0.31 |
Diras2 |
DIRAS family, GTP-binding RAS-like 2 |
1022 |
0.66 |
| chr9_72925072_72926177 | 0.31 |
Pygo1 |
pygopus 1 |
21 |
0.95 |
| chrX_166238673_166239964 | 0.31 |
Gpm6b |
glycoprotein m6b |
346 |
0.9 |
| chr1_157243489_157244692 | 0.30 |
Rasal2 |
RAS protein activator like 2 |
400 |
0.88 |
| chr10_78746927_78747974 | 0.30 |
Gm30400 |
predicted gene, 30400 |
291 |
0.84 |
| chr15_75746536_75747326 | 0.29 |
Mafa |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
1297 |
0.33 |
| chr5_111419755_111420664 | 0.29 |
Mn1 |
meningioma 1 |
2767 |
0.22 |
| chr2_28195247_28196182 | 0.29 |
Olfm1 |
olfactomedin 1 |
2621 |
0.32 |
| chr10_80226705_80227856 | 0.29 |
Pwwp3a |
PWWP domain containing 3A, DNA repair factor |
529 |
0.58 |
| chr7_99468579_99468981 | 0.29 |
Klhl35 |
kelch-like 35 |
183 |
0.9 |
| chr7_46179165_46180456 | 0.29 |
Abcc8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
176 |
0.92 |
| chr2_25567077_25567419 | 0.28 |
Mamdc4 |
MAM domain containing 4 |
1867 |
0.13 |
| chr7_126692598_126692845 | 0.28 |
Bola2 |
bolA-like 2 (E. coli) |
2680 |
0.09 |
| chr4_62499250_62499401 | 0.28 |
Hdhd3 |
haloacid dehalogenase-like hydrolase domain containing 3 |
2951 |
0.15 |
| chr1_34579671_34580338 | 0.28 |
1700101I19Rik |
RIKEN cDNA 1700101I19 gene |
293 |
0.5 |
| chr7_112225522_112225723 | 0.28 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
234 |
0.96 |
| chr7_141221857_141222428 | 0.28 |
Mir210 |
microRNA 210 |
649 |
0.44 |
| chr10_41070697_41071486 | 0.28 |
Gpr6 |
G protein-coupled receptor 6 |
1194 |
0.41 |
| chr18_40256495_40257230 | 0.28 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
100 |
0.96 |
| chr13_18948071_18948332 | 0.27 |
Amph |
amphiphysin |
4 |
0.97 |
| chr12_112779696_112780559 | 0.27 |
Ahnak2 |
AHNAK nucleoprotein 2 |
884 |
0.42 |
| chr1_180894021_180894978 | 0.27 |
Lefty2 |
left-right determination factor 2 |
41 |
0.95 |
| chr5_36529349_36530504 | 0.27 |
Tbc1d14 |
TBC1 domain family, member 14 |
581 |
0.71 |
| chr9_53705377_53706662 | 0.26 |
Rab39 |
RAB39, member RAS oncogene family |
213 |
0.91 |
| chr19_45228695_45230660 | 0.26 |
Lbx1 |
ladybird homeobox 1 |
6135 |
0.2 |
| chr5_73006697_73007924 | 0.25 |
Slc10a4 |
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
407 |
0.74 |
| chr7_5052519_5053499 | 0.25 |
Zfp580 |
zinc finger protein 580 |
1020 |
0.21 |
| chr15_76517568_76519917 | 0.25 |
Scrt1 |
scratch family zinc finger 1 |
3160 |
0.09 |
| chr6_128842519_128843166 | 0.25 |
Gm44066 |
predicted gene, 44066 |
329 |
0.76 |
| chr4_134747576_134747727 | 0.25 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
2456 |
0.3 |
| chr1_181352204_181352681 | 0.25 |
Cnih3 |
cornichon family AMPA receptor auxiliary protein 3 |
186 |
0.94 |
| chr6_39256378_39257582 | 0.24 |
Gm26008 |
predicted gene, 26008 |
8962 |
0.16 |
| chr14_70079421_70080436 | 0.24 |
Egr3 |
early growth response 3 |
1189 |
0.41 |
| chr6_29272372_29272690 | 0.24 |
Hilpda |
hypoxia inducible lipid droplet associated |
43 |
0.96 |
| chr2_112239763_112240639 | 0.24 |
Lpcat4 |
lysophosphatidylcholine acyltransferase 4 |
733 |
0.53 |
| chr2_25706786_25707496 | 0.24 |
Bmyc |
brain expressed myelocytomatosis oncogene |
402 |
0.61 |
| chr2_116790960_116791361 | 0.24 |
Gm13990 |
predicted gene 13990 |
86008 |
0.08 |
| chr8_78434733_78435587 | 0.23 |
Pou4f2 |
POU domain, class 4, transcription factor 2 |
1485 |
0.43 |
| chr17_43629214_43630519 | 0.23 |
Tdrd6 |
tudor domain containing 6 |
400 |
0.83 |
| chr1_93100315_93101980 | 0.23 |
Kif1a |
kinesin family member 1A |
675 |
0.62 |
| chr11_116653200_116654630 | 0.23 |
Prcd |
photoreceptor disc component |
60 |
0.83 |
| chr7_35554882_35556130 | 0.23 |
B230322F03Rik |
RIKEN cDNA B230322F03 gene |
139 |
0.71 |
| chr18_6765228_6765784 | 0.23 |
Rab18 |
RAB18, member RAS oncogene family |
286 |
0.9 |
| chr11_43835659_43836527 | 0.23 |
Adra1b |
adrenergic receptor, alpha 1b |
239 |
0.95 |
| chr11_98774111_98775335 | 0.23 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
610 |
0.56 |
| chr15_98792393_98793738 | 0.22 |
Wnt1 |
wingless-type MMTV integration site family, member 1 |
3208 |
0.11 |
| chr14_5070665_5071726 | 0.22 |
Gm8281 |
predicted gene, 8281 |
155 |
0.94 |
| chr4_106900804_106900955 | 0.22 |
Ssbp3 |
single-stranded DNA binding protein 3 |
9822 |
0.2 |
| chr4_76453825_76453976 | 0.22 |
Gm42303 |
predicted gene, 42303 |
2669 |
0.3 |
| chr7_4520460_4522358 | 0.22 |
Tnni3 |
troponin I, cardiac 3 |
842 |
0.35 |
| chr9_89910732_89911242 | 0.22 |
Rasgrf1 |
RAS protein-specific guanine nucleotide-releasing factor 1 |
1075 |
0.57 |
| chr2_29195751_29195902 | 0.22 |
Ntng2 |
netrin G2 |
8198 |
0.19 |
| chr7_140899782_140900558 | 0.22 |
Cox8b |
cytochrome c oxidase subunit 8B |
276 |
0.74 |
| chr1_78016726_78017156 | 0.22 |
Gm28387 |
predicted gene 28387 |
44565 |
0.18 |
| chr17_25836281_25837614 | 0.21 |
Rhbdl1 |
rhomboid like 1 |
135 |
0.84 |
| chr13_32338049_32338600 | 0.21 |
Gmds |
GDP-mannose 4, 6-dehydratase |
261 |
0.71 |
| chr11_116840114_116840538 | 0.21 |
Jmjd6 |
jumonji domain containing 6 |
925 |
0.4 |
| chr6_145933944_145934978 | 0.21 |
Sspn |
sarcospan |
339 |
0.86 |
| chr7_100371109_100371874 | 0.21 |
Ppme1 |
protein phosphatase methylesterase 1 |
443 |
0.59 |
| chr7_130626987_130627138 | 0.21 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
18419 |
0.16 |
| chr19_5039775_5039987 | 0.20 |
B4gat1 |
beta-1,4-glucuronyltransferase 1 |
974 |
0.21 |
| chr9_107399475_107400943 | 0.20 |
Cacna2d2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
140 |
0.93 |
| chr17_23993262_23994215 | 0.20 |
Prss22 |
protease, serine 22 |
4362 |
0.08 |
| chrX_74328789_74330084 | 0.20 |
Plxna3 |
plexin A3 |
370 |
0.67 |
| chr12_112784284_112784667 | 0.20 |
Ahnak2 |
AHNAK nucleoprotein 2 |
3464 |
0.13 |
| chr3_41564925_41565593 | 0.20 |
Jade1 |
jade family PHD finger 1 |
339 |
0.82 |
| chr11_96274202_96274759 | 0.20 |
Hoxb9 |
homeobox B9 |
2957 |
0.1 |
| chr18_34005890_34007297 | 0.20 |
Epb41l4a |
erythrocyte membrane protein band 4.1 like 4a |
326 |
0.88 |
| chr8_124426147_124426298 | 0.20 |
Pgbd5 |
piggyBac transposable element derived 5 |
7714 |
0.21 |
| chr9_50861540_50861809 | 0.20 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
4695 |
0.18 |
| chr15_76737177_76737328 | 0.20 |
Arhgap39 |
Rho GTPase activating protein 39 |
11649 |
0.08 |
| chr2_162661151_162661382 | 0.20 |
9430021M05Rik |
RIKEN cDNA 9430021M05 gene |
103 |
0.52 |
| chr8_87472820_87473580 | 0.19 |
Gm2694 |
predicted gene 2694 |
43 |
0.9 |
| chr7_69492924_69493481 | 0.19 |
Gm38584 |
predicted gene, 38584 |
2401 |
0.24 |
| chr3_55055185_55055856 | 0.19 |
Ccna1 |
cyclin A1 |
19 |
0.79 |
| chr13_46670045_46670243 | 0.19 |
Fam8a1 |
family with sequence similarity 8, member A1 |
622 |
0.69 |
| chr14_79769842_79771877 | 0.19 |
Pcdh8 |
protocadherin 8 |
345 |
0.74 |
| chr15_60822830_60824874 | 0.19 |
9930014A18Rik |
RIKEN cDNA 9930014A18 gene |
450 |
0.73 |
| chr15_98167479_98168718 | 0.19 |
Ccdc184 |
coiled-coil domain containing 184 |
940 |
0.4 |
| chr5_71657120_71658579 | 0.18 |
Gabra4 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
26 |
0.79 |
| chr12_112781818_112782096 | 0.18 |
Ahnak2 |
AHNAK nucleoprotein 2 |
946 |
0.39 |
| chr8_24674733_24674884 | 0.18 |
Adam18 |
a disintegrin and metallopeptidase domain 18 |
53 |
0.97 |
| chr6_50054234_50054467 | 0.18 |
Gm3455 |
predicted gene 3455 |
3164 |
0.33 |
| chr4_49844475_49845850 | 0.18 |
Grin3a |
glutamate receptor ionotropic, NMDA3A |
387 |
0.91 |
| chr5_110543976_110545228 | 0.18 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
247 |
0.9 |
| chr17_35059046_35060516 | 0.18 |
Ddah2 |
dimethylarginine dimethylaminohydrolase 2 |
35 |
0.89 |
| chr1_130877918_130878766 | 0.18 |
Fcmr |
Fc fragment of IgM receptor |
11 |
0.97 |
| chr2_13010530_13011322 | 0.18 |
C1ql3 |
C1q-like 3 |
880 |
0.34 |
| chr11_118570728_118570912 | 0.18 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
910 |
0.64 |
| chr11_49168954_49169714 | 0.18 |
Btnl9 |
butyrophilin-like 9 |
1341 |
0.28 |
| chr12_112783087_112783238 | 0.17 |
Ahnak2 |
AHNAK nucleoprotein 2 |
2151 |
0.18 |
| chr4_146058803_146058954 | 0.17 |
Gm13036 |
predicted gene 13036 |
102 |
0.96 |
| chr4_153484340_153484575 | 0.17 |
Ajap1 |
adherens junction associated protein 1 |
1646 |
0.54 |
| chr7_4835870_4836475 | 0.17 |
Shisa7 |
shisa family member 7 |
2 |
0.94 |
| chr7_79148061_79149306 | 0.17 |
Mfge8 |
milk fat globule-EGF factor 8 protein |
306 |
0.89 |
| chr14_63743158_63744317 | 0.17 |
Mir598 |
microRNA 598 |
16548 |
0.21 |
| chr2_120063894_120064045 | 0.17 |
Sptbn5 |
spectrin beta, non-erythrocytic 5 |
21709 |
0.1 |
| chr13_25269167_25270348 | 0.17 |
Nrsn1 |
neurensin 1 |
213 |
0.96 |
| chr7_4692884_4693035 | 0.17 |
Brsk1 |
BR serine/threonine kinase 1 |
716 |
0.43 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.7 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.3 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.4 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.8 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.1 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.2 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.5 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 0.1 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.3 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.5 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 1.1 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.0 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.0 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.8 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0043731 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.7 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0019176 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |