Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
Results for Hlx
Z-value: 0.59
Transcription factors associated with Hlx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlx
|
ENSMUSG00000039377.6 | H2.0-like homeobox |
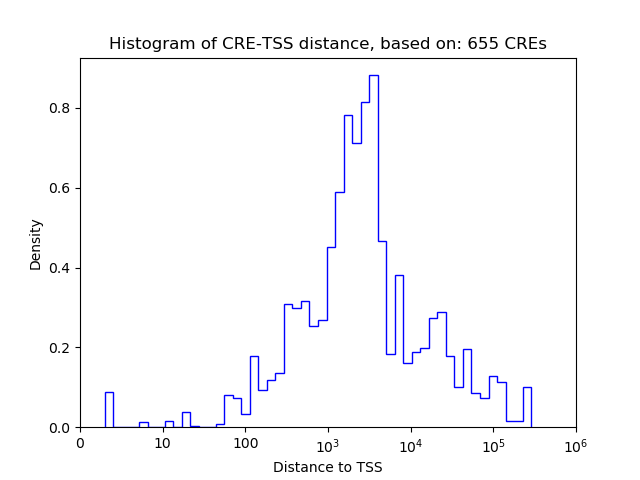
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_184733855_184734791 | Hlx | 1704 | 0.268399 | -0.61 | 2.9e-07 | Click! |
| chr1_184731287_184732512 | Hlx | 301 | 0.862596 | -0.54 | 7.9e-06 | Click! |
| chr1_184732758_184733253 | Hlx | 386 | 0.805138 | -0.49 | 6.2e-05 | Click! |
| chr1_184729496_184731200 | Hlx | 1250 | 0.373544 | -0.44 | 4.1e-04 | Click! |
| chr1_184728502_184729126 | Hlx | 2784 | 0.197330 | -0.43 | 7.1e-04 | Click! |
Activity of the Hlx motif across conditions
Conditions sorted by the z-value of the Hlx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
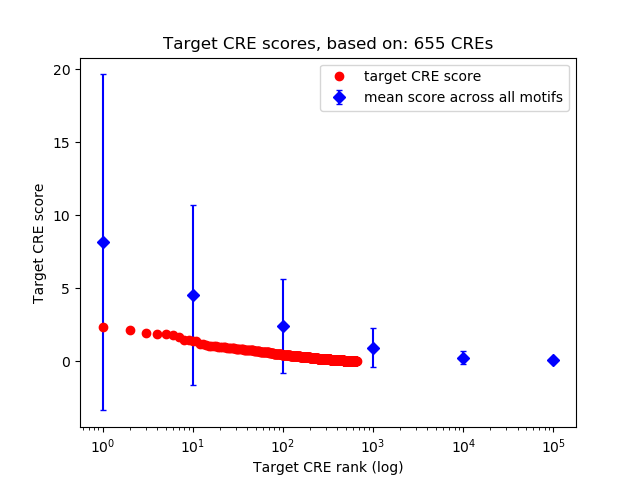
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_70842167_70842810 | 2.36 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
129 |
0.98 |
| chr5_120426818_120428597 | 2.14 |
Lhx5 |
LIM homeobox protein 5 |
3992 |
0.15 |
| chr7_87586513_87587584 | 1.91 |
Grm5 |
glutamate receptor, metabotropic 5 |
2650 |
0.4 |
| chr18_43686213_43686392 | 1.88 |
Jakmip2 |
janus kinase and microtubule interacting protein 2 |
1323 |
0.49 |
| chr13_69734884_69735178 | 1.85 |
Ube2ql1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
4858 |
0.14 |
| chr14_66865047_66865736 | 1.83 |
Dpysl2 |
dihydropyrimidinase-like 2 |
3297 |
0.19 |
| chr3_8512495_8512918 | 1.63 |
Stmn2 |
stathmin-like 2 |
3120 |
0.28 |
| chr14_55056074_55056891 | 1.48 |
Gm20687 |
predicted gene 20687 |
989 |
0.3 |
| chr1_169745785_169746010 | 1.47 |
Rgs4 |
regulator of G-protein signaling 4 |
1726 |
0.41 |
| chr15_103058659_103059955 | 1.40 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
1 |
0.95 |
| chr9_52148115_52149635 | 1.36 |
Zc3h12c |
zinc finger CCCH type containing 12C |
19236 |
0.18 |
| chr8_34890130_34891317 | 1.19 |
Tnks |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
572 |
0.8 |
| chr18_59062200_59063436 | 1.19 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
307 |
0.94 |
| chr3_38894285_38895428 | 1.11 |
Fat4 |
FAT atypical cadherin 4 |
3914 |
0.27 |
| chr3_89521563_89522618 | 1.08 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
1926 |
0.28 |
| chr3_63961199_63961587 | 1.08 |
Gm26850 |
predicted gene, 26850 |
2240 |
0.21 |
| chr18_54719777_54720132 | 1.07 |
Gm5821 |
predicted gene 5821 |
46178 |
0.16 |
| chr1_177448882_177449429 | 1.04 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
3334 |
0.19 |
| chr18_25745414_25746450 | 1.00 |
Celf4 |
CUGBP, Elav-like family member 4 |
6760 |
0.24 |
| chr2_123533004_123533690 | 1.00 |
Gm13988 |
predicted gene 13988 |
259423 |
0.02 |
| chr9_41585694_41587243 | 0.97 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
1301 |
0.29 |
| chr3_134236641_134237783 | 0.97 |
Cxxc4 |
CXXC finger 4 |
392 |
0.78 |
| chr7_123984527_123984925 | 0.96 |
Gm27040 |
predicted gene, 27040 |
1272 |
0.38 |
| chr3_86748624_86748824 | 0.93 |
Gm37876 |
predicted gene, 37876 |
29929 |
0.17 |
| chr7_137305711_137306880 | 0.92 |
Ebf3 |
early B cell factor 3 |
7621 |
0.2 |
| chr8_78433398_78433999 | 0.90 |
Pou4f2 |
POU domain, class 4, transcription factor 2 |
2947 |
0.27 |
| chr14_108912235_108913525 | 0.89 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
1278 |
0.64 |
| chr1_9296437_9296701 | 0.89 |
Sntg1 |
syntrophin, gamma 1 |
1667 |
0.35 |
| chr10_69536775_69537233 | 0.89 |
Ank3 |
ankyrin 3, epithelial |
2782 |
0.32 |
| chr5_135248496_135249658 | 0.87 |
Fzd9 |
frizzled class receptor 9 |
2153 |
0.21 |
| chr11_32001099_32002296 | 0.87 |
Nsg2 |
neuron specific gene family member 2 |
1195 |
0.52 |
| chr2_146061763_146063020 | 0.86 |
Cfap61 |
cilia and flagella associated protein 61 |
15140 |
0.25 |
| chr2_151631540_151632560 | 0.85 |
Snph |
syntaphilin |
421 |
0.78 |
| chr1_115688015_115688174 | 0.85 |
Cntnap5a |
contactin associated protein-like 5A |
3338 |
0.29 |
| chr1_136228373_136230942 | 0.84 |
Inava |
innate immunity activator |
362 |
0.76 |
| chr13_104111586_104112312 | 0.81 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
2120 |
0.26 |
| chr4_33926104_33927188 | 0.81 |
Cnr1 |
cannabinoid receptor 1 (brain) |
444 |
0.88 |
| chr12_31711352_31711698 | 0.80 |
Gpr22 |
G protein-coupled receptor 22 |
2401 |
0.25 |
| chr5_37241461_37244349 | 0.78 |
Crmp1 |
collapsin response mediator protein 1 |
171 |
0.95 |
| chr19_44748114_44748265 | 0.78 |
Gm35610 |
predicted gene, 35610 |
6836 |
0.15 |
| chr7_79504311_79505700 | 0.78 |
Mir9-3 |
microRNA 9-3 |
259 |
0.82 |
| chr18_37217058_37218378 | 0.78 |
Gm10544 |
predicted gene 10544 |
39196 |
0.08 |
| chr6_112609185_112609336 | 0.76 |
Gm5578 |
predicted pseudogene 5578 |
3716 |
0.21 |
| chr7_99275133_99275418 | 0.76 |
Map6 |
microtubule-associated protein 6 |
6143 |
0.13 |
| chr19_20009817_20010437 | 0.75 |
Gm22684 |
predicted gene, 22684 |
23508 |
0.22 |
| chr13_83736071_83736534 | 0.75 |
Gm33366 |
predicted gene, 33366 |
2233 |
0.18 |
| chr11_23893045_23893724 | 0.74 |
Gm12061 |
predicted gene 12061 |
1176 |
0.38 |
| chr15_85682337_85682535 | 0.72 |
Lncppara |
long noncoding RNA near Ppara |
21337 |
0.12 |
| chr3_4796861_4798079 | 0.71 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
88 |
0.98 |
| chr12_79814152_79814328 | 0.71 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
110493 |
0.06 |
| chr2_163977230_163977381 | 0.71 |
Ywhab |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
17655 |
0.16 |
| chr4_99659246_99659665 | 0.70 |
Gm23366 |
predicted gene, 23366 |
1713 |
0.26 |
| chr9_56159774_56159955 | 0.70 |
Tspan3 |
tetraspanin 3 |
678 |
0.68 |
| chr14_104641438_104641957 | 0.69 |
D130009I18Rik |
RIKEN cDNA D130009I18 gene |
2553 |
0.3 |
| chrX_6171274_6171425 | 0.69 |
Nudt10 |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
1666 |
0.4 |
| chr12_117345149_117346019 | 0.67 |
Gm5441 |
predicted gene 5441 |
8562 |
0.29 |
| chr10_94941542_94941964 | 0.66 |
Plxnc1 |
plexin C1 |
3082 |
0.28 |
| chr11_96002027_96002959 | 0.66 |
Gm29202 |
predicted gene 29202 |
3396 |
0.11 |
| chr11_111605019_111605670 | 0.65 |
Gm11676 |
predicted gene 11676 |
7962 |
0.32 |
| chr13_20472048_20472724 | 0.64 |
Elmo1 |
engulfment and cell motility 1 |
340 |
0.8 |
| chr3_34654574_34655689 | 0.64 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
905 |
0.42 |
| chr19_12498310_12498475 | 0.64 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
3062 |
0.14 |
| chr3_119154592_119155474 | 0.64 |
Gm43410 |
predicted gene 43410 |
286789 |
0.01 |
| chr14_104463807_104464161 | 0.64 |
Pou4f1 |
POU domain, class 4, transcription factor 1 |
1381 |
0.4 |
| chr1_19213854_19215338 | 0.63 |
Tfap2b |
transcription factor AP-2 beta |
717 |
0.69 |
| chr14_64233514_64233974 | 0.63 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
117430 |
0.05 |
| chr1_172341079_172341970 | 0.63 |
Kcnj10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
314 |
0.81 |
| chr6_7554855_7556232 | 0.62 |
Tac1 |
tachykinin 1 |
447 |
0.85 |
| chr12_46813712_46814094 | 0.62 |
Gm48542 |
predicted gene, 48542 |
2395 |
0.29 |
| chr19_47018258_47018719 | 0.60 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
3335 |
0.15 |
| chr7_51629095_51630495 | 0.58 |
Slc17a6 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
315 |
0.88 |
| chr2_65620767_65621991 | 0.57 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
568 |
0.82 |
| chr8_110275623_110275994 | 0.56 |
Gm26832 |
predicted gene, 26832 |
8346 |
0.19 |
| chr15_92598307_92599654 | 0.56 |
Pdzrn4 |
PDZ domain containing RING finger 4 |
1854 |
0.49 |
| chr9_74869680_74869831 | 0.55 |
Onecut1 |
one cut domain, family member 1 |
3271 |
0.2 |
| chr9_75681964_75682559 | 0.55 |
Scg3 |
secretogranin III |
1326 |
0.37 |
| chr2_152080491_152081480 | 0.54 |
Scrt2 |
scratch family zinc finger 2 |
544 |
0.7 |
| chr5_63651264_63652181 | 0.54 |
Gm9954 |
predicted gene 9954 |
828 |
0.61 |
| chr1_15288505_15288656 | 0.53 |
Kcnb2 |
potassium voltage gated channel, Shab-related subfamily, member 2 |
1326 |
0.51 |
| chr8_41052368_41053980 | 0.53 |
Gm16193 |
predicted gene 16193 |
64 |
0.96 |
| chr14_122481884_122483266 | 0.53 |
Zic2 |
zinc finger protein of the cerebellum 2 |
4475 |
0.12 |
| chr2_116073034_116073185 | 0.52 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
2987 |
0.22 |
| chr18_72349127_72350542 | 0.52 |
Dcc |
deleted in colorectal carcinoma |
1183 |
0.64 |
| chr3_86543379_86544222 | 0.51 |
Lrba |
LPS-responsive beige-like anchor |
1767 |
0.38 |
| chr3_131341006_131341355 | 0.51 |
Sgms2 |
sphingomyelin synthase 2 |
3753 |
0.2 |
| chr10_89875205_89875398 | 0.50 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
1629 |
0.45 |
| chr17_67499984_67500234 | 0.50 |
Gm36201 |
predicted gene, 36201 |
129223 |
0.05 |
| chr12_52700044_52701597 | 0.50 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
1437 |
0.46 |
| chr15_18819282_18819740 | 0.50 |
Cdh10 |
cadherin 10 |
534 |
0.58 |
| chr7_35848412_35848964 | 0.49 |
Gm28514 |
predicted gene 28514 |
10428 |
0.21 |
| chr11_96593909_96594964 | 0.48 |
Skap1 |
src family associated phosphoprotein 1 |
104575 |
0.05 |
| chr4_128885478_128885629 | 0.48 |
Trim62 |
tripartite motif-containing 62 |
1965 |
0.27 |
| chr10_90578974_90579573 | 0.48 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
2281 |
0.42 |
| chr10_90577565_90578158 | 0.47 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
869 |
0.72 |
| chrX_153501207_153502250 | 0.47 |
Ubqln2 |
ubiquilin 2 |
3501 |
0.22 |
| chr2_72426765_72427714 | 0.47 |
Cdca7 |
cell division cycle associated 7 |
48920 |
0.13 |
| chr8_47345612_47345906 | 0.46 |
Stox2 |
storkhead box 2 |
6589 |
0.26 |
| chr11_84520959_84524590 | 0.46 |
Lhx1 |
LIM homeobox protein 1 |
63 |
0.97 |
| chr18_42429204_42429681 | 0.46 |
Gm16415 |
predicted pseudogene 16415 |
11577 |
0.18 |
| chr4_124254790_124255065 | 0.46 |
Gm37667 |
predicted gene, 37667 |
24250 |
0.16 |
| chr9_91404809_91406365 | 0.45 |
Gm29478 |
predicted gene 29478 |
1113 |
0.42 |
| chr17_56241459_56242409 | 0.45 |
A230051N06Rik |
RIKEN cDNA A230051N06 gene |
218 |
0.81 |
| chr10_45889498_45890055 | 0.44 |
Gpx4-ps2 |
glutathione peroxidase 4, pseudogene 2 |
14471 |
0.23 |
| chr7_70355803_70356454 | 0.43 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
891 |
0.46 |
| chr10_87500739_87501897 | 0.43 |
Gm48120 |
predicted gene, 48120 |
6544 |
0.19 |
| chr9_72533965_72534704 | 0.43 |
Rfx7 |
regulatory factor X, 7 |
1599 |
0.21 |
| chr16_38093809_38093980 | 0.43 |
Gsk3b |
glycogen synthase kinase 3 beta |
3606 |
0.27 |
| chr16_74408614_74409150 | 0.43 |
Robo2 |
roundabout guidance receptor 2 |
2030 |
0.43 |
| chr13_84905027_84905302 | 0.42 |
Gm4059 |
predicted gene 4059 |
69153 |
0.12 |
| chr2_14740186_14740953 | 0.42 |
Gm10848 |
predicted gene 10848 |
847 |
0.36 |
| chr16_28751716_28751977 | 0.42 |
Fgf12 |
fibroblast growth factor 12 |
1222 |
0.62 |
| chr1_153665136_153666782 | 0.42 |
Rgs8 |
regulator of G-protein signaling 8 |
250 |
0.89 |
| chrX_146965194_146965345 | 0.42 |
Htr2c |
5-hydroxytryptamine (serotonin) receptor 2C |
2246 |
0.23 |
| chr8_106985656_106985807 | 0.42 |
Gm22085 |
predicted gene, 22085 |
23880 |
0.1 |
| chr8_108716860_108718878 | 0.41 |
Zfhx3 |
zinc finger homeobox 3 |
3225 |
0.3 |
| chr2_73267880_73268204 | 0.41 |
Sp9 |
trans-acting transcription factor 9 |
3883 |
0.19 |
| chr2_79047843_79048374 | 0.41 |
Gm14469 |
predicted gene 14469 |
9374 |
0.22 |
| chrX_166344665_166345995 | 0.40 |
Gpm6b |
glycoprotein m6b |
488 |
0.85 |
| chr17_45570526_45571346 | 0.40 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
646 |
0.5 |
| chr14_7821774_7821925 | 0.40 |
Flnb |
filamin, beta |
3892 |
0.19 |
| chr4_103619552_103620735 | 0.39 |
Dab1 |
disabled 1 |
478 |
0.8 |
| chr7_70363361_70364056 | 0.39 |
B130024G19Rik |
RIKEN cDNA B130024G19 gene |
1202 |
0.34 |
| chr14_32601725_32601876 | 0.39 |
Prrxl1 |
paired related homeobox protein-like 1 |
1842 |
0.31 |
| chr7_35949147_35949298 | 0.39 |
Gm28514 |
predicted gene 28514 |
110962 |
0.06 |
| chr7_36703759_36704512 | 0.38 |
Tshz3 |
teashirt zinc finger family member 3 |
5918 |
0.14 |
| chr3_80799469_80799764 | 0.38 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
2963 |
0.34 |
| chr15_102984578_102985289 | 0.38 |
Hoxc8 |
homeobox C8 |
5674 |
0.08 |
| chr11_24085127_24085687 | 0.38 |
Bcl11a |
B cell CLL/lymphoma 11A (zinc finger protein) |
4737 |
0.16 |
| chr2_158595089_158595667 | 0.38 |
Gm14204 |
predicted gene 14204 |
15212 |
0.11 |
| chr7_70365047_70366578 | 0.38 |
B130024G19Rik |
RIKEN cDNA B130024G19 gene |
653 |
0.47 |
| chr2_74695738_74695889 | 0.38 |
Gm14396 |
predicted gene 14396 |
212 |
0.75 |
| chr3_31099281_31100546 | 0.37 |
Skil |
SKI-like |
3080 |
0.26 |
| chr2_38341551_38341924 | 0.37 |
Lhx2 |
LIM homeobox protein 2 |
645 |
0.64 |
| chr13_36728088_36728779 | 0.37 |
Gm30177 |
predicted gene, 30177 |
1175 |
0.4 |
| chr15_99056560_99057587 | 0.37 |
Prph |
peripherin |
1103 |
0.3 |
| chr3_7505053_7505454 | 0.37 |
Zc2hc1a |
zinc finger, C2HC-type containing 1A |
1767 |
0.33 |
| chr17_91085493_91086001 | 0.37 |
Gm47307 |
predicted gene, 47307 |
2659 |
0.21 |
| chr4_24429901_24430719 | 0.36 |
Gm27243 |
predicted gene 27243 |
580 |
0.79 |
| chr14_61600038_61600691 | 0.36 |
Trim13 |
tripartite motif-containing 13 |
824 |
0.41 |
| chrX_58027481_58028617 | 0.36 |
Zic3 |
zinc finger protein of the cerebellum 3 |
2594 |
0.35 |
| chr14_122344326_122345026 | 0.36 |
Gm25464 |
predicted gene, 25464 |
43484 |
0.12 |
| chr5_13125231_13126281 | 0.35 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
163 |
0.95 |
| chr16_77852573_77852735 | 0.35 |
Gm17333 |
predicted gene, 17333 |
6050 |
0.27 |
| chr15_102593847_102594169 | 0.34 |
Atf7 |
activating transcription factor 7 |
30914 |
0.08 |
| chr15_59876437_59876598 | 0.34 |
Gm7708 |
predicted gene 7708 |
53773 |
0.13 |
| chr16_80017098_80017433 | 0.34 |
1700066C05Rik |
RIKEN cDNA 1700066C05 gene |
17889 |
0.27 |
| chr13_63627159_63628101 | 0.34 |
Gm30709 |
predicted gene, 30709 |
251 |
0.9 |
| chr7_124990750_124991105 | 0.34 |
Gm45093 |
predicted gene 45093 |
78564 |
0.09 |
| chr5_118169234_118170295 | 0.34 |
Hrk |
harakiri, BCL2 interacting protein (contains only BH3 domain) |
16 |
0.97 |
| chr6_15196934_15197697 | 0.34 |
Foxp2 |
forkhead box P2 |
351 |
0.94 |
| chr2_165367693_165368982 | 0.33 |
Zfp663 |
zinc finger protein 663 |
386 |
0.8 |
| chr11_50222884_50223393 | 0.33 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2197 |
0.17 |
| chr4_45824039_45824548 | 0.33 |
Igfbpl1 |
insulin-like growth factor binding protein-like 1 |
2630 |
0.22 |
| chr10_127421987_127422298 | 0.33 |
R3hdm2 |
R3H domain containing 2 |
910 |
0.44 |
| chr5_13127003_13127271 | 0.33 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
1544 |
0.35 |
| chr1_78172409_78172663 | 0.33 |
Pax3 |
paired box 3 |
24302 |
0.21 |
| chr4_141734679_141735016 | 0.32 |
Ddi2 |
DNA-damage inducible protein 2 |
11428 |
0.12 |
| chr14_24002097_24002465 | 0.32 |
4930519K11Rik |
RIKEN cDNA 4930519K11 gene |
1563 |
0.3 |
| chr14_84450287_84451113 | 0.32 |
Pcdh17 |
protocadherin 17 |
2193 |
0.37 |
| chr7_73554401_73554784 | 0.32 |
1810026B05Rik |
RIKEN cDNA 1810026B05 gene |
1328 |
0.3 |
| chr7_73151898_73152049 | 0.32 |
4933435G04Rik |
RIKEN cDNA 4933435G04 gene |
27522 |
0.13 |
| chr14_64594009_64594633 | 0.32 |
Mir3078 |
microRNA 3078 |
3136 |
0.2 |
| chr12_109543555_109544268 | 0.32 |
Meg3 |
maternally expressed 3 |
587 |
0.28 |
| chr1_12991432_12992703 | 0.32 |
Slco5a1 |
solute carrier organic anion transporter family, member 5A1 |
583 |
0.78 |
| chr2_116074186_116074465 | 0.31 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
1771 |
0.31 |
| chr6_101194590_101194876 | 0.31 |
Gm26911 |
predicted gene, 26911 |
3836 |
0.2 |
| chr17_83792570_83792796 | 0.31 |
Mta3 |
metastasis associated 3 |
2739 |
0.29 |
| chr7_137311015_137311991 | 0.30 |
Ebf3 |
early B cell factor 3 |
2413 |
0.29 |
| chr15_36967150_36968398 | 0.30 |
Gm34590 |
predicted gene, 34590 |
28910 |
0.13 |
| chr11_111522983_111523181 | 0.30 |
Gm11676 |
predicted gene 11676 |
90224 |
0.1 |
| chr3_5212584_5213216 | 0.30 |
Gm10748 |
predicted gene 10748 |
62 |
0.97 |
| chr11_60182522_60182943 | 0.30 |
Rai1 |
retinoic acid induced 1 |
6853 |
0.13 |
| chr12_52435515_52435783 | 0.29 |
Gm47431 |
predicted gene, 47431 |
12476 |
0.21 |
| chr5_120304073_120304707 | 0.29 |
Gm26474 |
predicted gene, 26474 |
16664 |
0.17 |
| chr1_188065284_188065435 | 0.29 |
9330162B11Rik |
RIKEN cDNA 9330162B11 gene |
56369 |
0.14 |
| chrX_64273727_64274331 | 0.28 |
Slitrk4 |
SLIT and NTRK-like family, member 4 |
1793 |
0.49 |
| chrX_133682515_133683917 | 0.28 |
Pcdh19 |
protocadherin 19 |
1775 |
0.49 |
| chr9_60798563_60798845 | 0.28 |
Uaca |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
4079 |
0.22 |
| chr17_51760240_51761547 | 0.28 |
C230085N15Rik |
RIKEN cDNA C230085N15 gene |
728 |
0.54 |
| chr3_97763340_97765161 | 0.28 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
3681 |
0.23 |
| chr1_155414403_155414717 | 0.28 |
Xpr1 |
xenotropic and polytropic retrovirus receptor 1 |
2769 |
0.34 |
| chr2_80126598_80127760 | 0.28 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
1655 |
0.42 |
| chr12_27339427_27339967 | 0.28 |
Sox11 |
SRY (sex determining region Y)-box 11 |
2877 |
0.38 |
| chr18_54125378_54126373 | 0.28 |
Gm8594 |
predicted gene 8594 |
94567 |
0.09 |
| chr7_49907312_49908741 | 0.28 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
2120 |
0.4 |
| chrX_38473570_38474329 | 0.28 |
Gm7598 |
predicted gene 7598 |
2512 |
0.27 |
| chr17_44810813_44811180 | 0.27 |
Runx2 |
runt related transcription factor 2 |
3230 |
0.26 |
| chr14_61139006_61139387 | 0.27 |
Sacs |
sacsin |
705 |
0.73 |
| chr14_72155158_72155309 | 0.27 |
Gm23735 |
predicted gene, 23735 |
24145 |
0.23 |
| chr3_63961758_63962185 | 0.27 |
Gm26850 |
predicted gene, 26850 |
1662 |
0.26 |
| chr7_87589934_87590085 | 0.27 |
Grm5 |
glutamate receptor, metabotropic 5 |
5611 |
0.31 |
| chr2_28837901_28838652 | 0.27 |
Gtf3c4 |
general transcription factor IIIC, polypeptide 4 |
2010 |
0.23 |
| chr9_79875436_79876355 | 0.26 |
Gm3211 |
predicted gene 3211 |
37305 |
0.12 |
| chr19_57236960_57237217 | 0.26 |
Ablim1 |
actin-binding LIM protein 1 |
2241 |
0.34 |
| chr5_127631930_127633243 | 0.26 |
Slc15a4 |
solute carrier family 15, member 4 |
294 |
0.51 |
| chr3_34196316_34197672 | 0.26 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
130 |
0.97 |
| chr5_137039973_137040124 | 0.26 |
Ap1s1 |
adaptor protein complex AP-1, sigma 1 |
2398 |
0.16 |
| chr19_7384327_7384630 | 0.26 |
Spindoc |
spindlin interactor and repressor of chromatin binding |
1484 |
0.22 |
| chr19_20010440_20010591 | 0.26 |
Gm22684 |
predicted gene, 22684 |
23120 |
0.22 |
| chr7_61309034_61309202 | 0.26 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
2595 |
0.4 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 0.8 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.2 | 0.5 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.2 | 0.5 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.5 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.4 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.9 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 2.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.5 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.1 | 0.4 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.1 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.5 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:2001258 | negative regulation of cation channel activity(GO:2001258) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.0 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 2.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.4 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |