Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
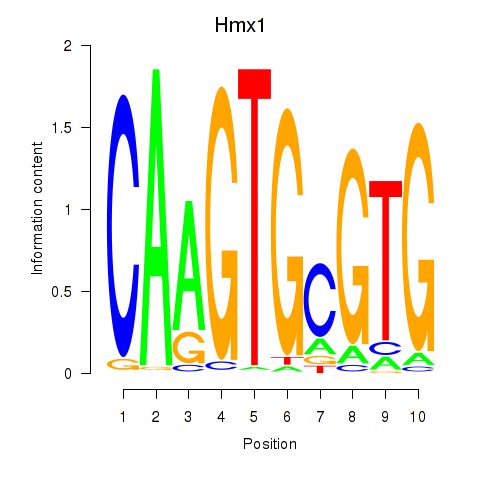
Results for Hmx1
Z-value: 0.62
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSMUSG00000067438.3 | H6 homeobox 1 |
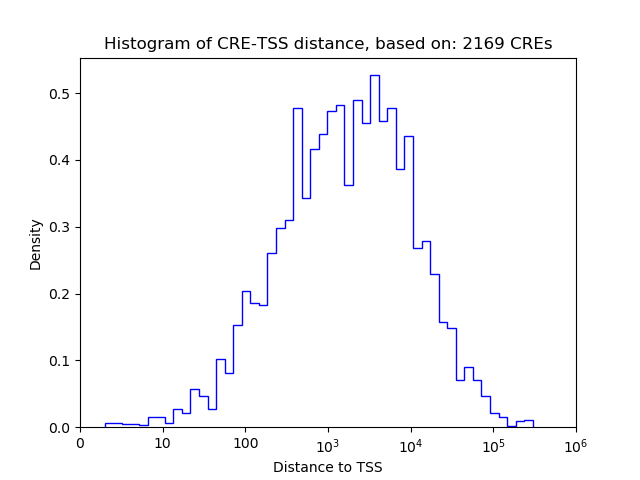
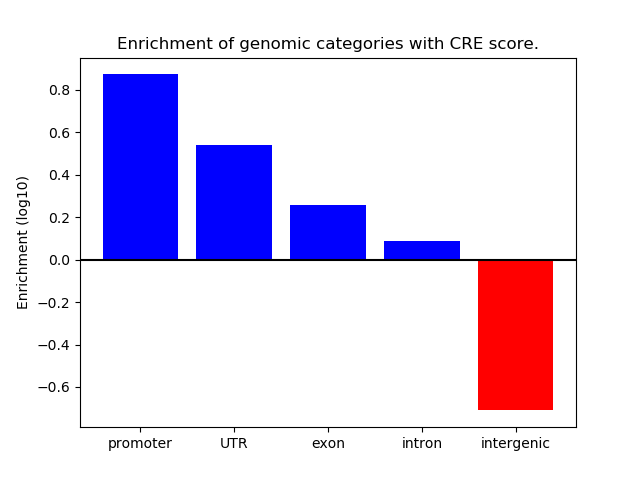
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_35388341_35389287 | Hmx1 | 294 | 0.512607 | -0.29 | 2.7e-02 | Click! |
| chr5_35389371_35389565 | Hmx1 | 351 | 0.725062 | -0.20 | 1.3e-01 | Click! |
| chr5_35389846_35390469 | Hmx1 | 1040 | 0.338614 | -0.12 | 3.7e-01 | Click! |
Activity of the Hmx1 motif across conditions
Conditions sorted by the z-value of the Hmx1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
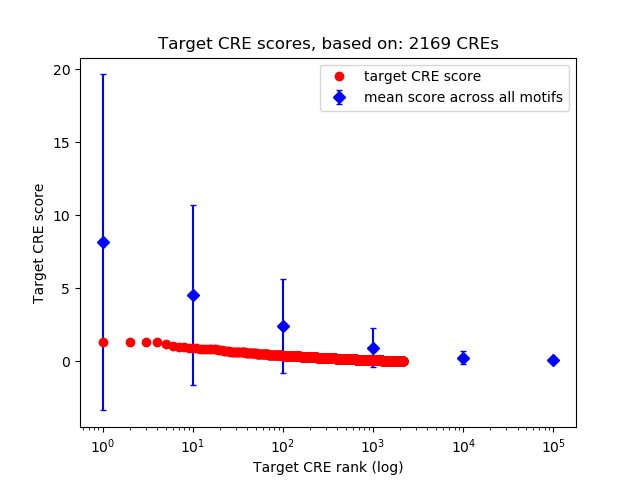
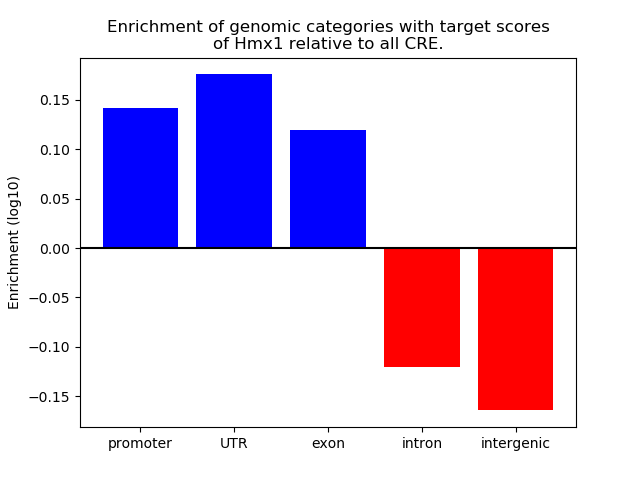
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_129216664_129219042 | 1.35 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
3578 |
0.2 |
| chr6_39494466_39494617 | 1.33 |
Dennd2a |
DENN/MADD domain containing 2A |
5375 |
0.17 |
| chr13_4233789_4234486 | 1.31 |
Akr1c19 |
aldo-keto reductase family 1, member C19 |
397 |
0.81 |
| chr7_120880267_120880570 | 1.30 |
Eef2k |
eukaryotic elongation factor-2 kinase |
4198 |
0.14 |
| chr11_96343236_96346574 | 1.19 |
Hoxb3 |
homeobox B3 |
1136 |
0.24 |
| chr2_158174901_158175285 | 1.07 |
1700060C20Rik |
RIKEN cDNA 1700060C20 gene |
16915 |
0.14 |
| chr7_132778272_132778793 | 0.95 |
Fam53b |
family with sequence similarity 53, member B |
1616 |
0.39 |
| chr8_121088119_121090419 | 0.95 |
Gm27530 |
predicted gene, 27530 |
4563 |
0.13 |
| chr19_58453572_58455406 | 0.94 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
23 |
0.98 |
| chr9_55282826_55283068 | 0.93 |
Nrg4 |
neuregulin 4 |
625 |
0.71 |
| chr17_48448887_48449311 | 0.92 |
Tspo2 |
translocator protein 2 |
971 |
0.43 |
| chr4_139336568_139337223 | 0.87 |
AL807811.1 |
aldo-keto reductase family 7 member A2 (AKR7A2) pseudogene |
285 |
0.73 |
| chr4_119188012_119188665 | 0.86 |
Ermap |
erythroblast membrane-associated protein |
409 |
0.69 |
| chr5_75075112_75077490 | 0.85 |
Gsx2 |
GS homeobox 2 |
699 |
0.56 |
| chr17_57227821_57228041 | 0.85 |
C3 |
complement component 3 |
72 |
0.95 |
| chr7_105762080_105763247 | 0.82 |
Gm15645 |
predicted gene 15645 |
342 |
0.76 |
| chr7_133700764_133701966 | 0.82 |
Uros |
uroporphyrinogen III synthase |
1173 |
0.35 |
| chr19_25505518_25507191 | 0.81 |
Dmrt1 |
doublesex and mab-3 related transcription factor 1 |
647 |
0.72 |
| chr12_103862299_103862565 | 0.79 |
Serpina1a |
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
1119 |
0.32 |
| chr12_103826543_103826975 | 0.76 |
Serpina1b |
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
3614 |
0.12 |
| chr2_167688724_167690153 | 0.75 |
Tmem189 |
transmembrane protein 189 |
384 |
0.52 |
| chr8_84703616_84705950 | 0.72 |
Nfix |
nuclear factor I/X |
2933 |
0.13 |
| chr12_84203794_84203995 | 0.72 |
Gm31513 |
predicted gene, 31513 |
7925 |
0.11 |
| chr11_58927689_58928812 | 0.68 |
Btnl10 |
butyrophilin-like 10 |
10193 |
0.06 |
| chr3_90368038_90368189 | 0.68 |
Gatad2b |
GATA zinc finger domain containing 2B |
16695 |
0.09 |
| chr13_91461118_91462460 | 0.67 |
Ssbp2 |
single-stranded DNA binding protein 2 |
608 |
0.81 |
| chr11_35951046_35951261 | 0.67 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
29374 |
0.19 |
| chr2_168617972_168618123 | 0.65 |
Nfatc2 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
16390 |
0.19 |
| chr8_119437065_119437967 | 0.65 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
327 |
0.86 |
| chr7_133114831_133116529 | 0.65 |
Ctbp2 |
C-terminal binding protein 2 |
2981 |
0.2 |
| chr3_37987146_37987297 | 0.65 |
Gm42751 |
predicted gene 42751 |
4429 |
0.19 |
| chr7_74421986_74422217 | 0.64 |
Gm7580 |
predicted gene 7580 |
29403 |
0.22 |
| chr15_31529901_31531020 | 0.62 |
Marchf6 |
membrane associated ring-CH-type finger 6 |
593 |
0.65 |
| chr8_46473091_46473536 | 0.62 |
Gm45244 |
predicted gene 45244 |
277 |
0.87 |
| chr19_10015065_10016667 | 0.62 |
Rab3il1 |
RAB3A interacting protein (rabin3)-like 1 |
822 |
0.48 |
| chr7_16416990_16417583 | 0.62 |
Gm45510 |
predicted gene 45510 |
665 |
0.51 |
| chr16_85797970_85798654 | 0.61 |
Adamts1 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
1326 |
0.51 |
| chr8_72665204_72665522 | 0.61 |
Nwd1 |
NACHT and WD repeat domain containing 1 |
8277 |
0.15 |
| chr15_80624523_80624695 | 0.60 |
Grap2 |
GRB2-related adaptor protein 2 |
1087 |
0.43 |
| chr6_138143280_138143517 | 0.59 |
Mgst1 |
microsomal glutathione S-transferase 1 |
544 |
0.85 |
| chr12_111833749_111833971 | 0.59 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
9095 |
0.11 |
| chr11_98766918_98767150 | 0.59 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3392 |
0.13 |
| chr19_6634111_6634262 | 0.59 |
Gm14968 |
predicted gene 14968 |
11501 |
0.14 |
| chr7_141654914_141655534 | 0.59 |
Muc6 |
mucin 6, gastric |
84 |
0.96 |
| chrX_53055829_53057122 | 0.58 |
C430049B03Rik |
RIKEN cDNA C430049B03 gene |
685 |
0.41 |
| chr19_10523211_10524258 | 0.58 |
Sdhaf2 |
succinate dehydrogenase complex assembly factor 2 |
1358 |
0.25 |
| chr5_64807638_64809344 | 0.56 |
Klf3 |
Kruppel-like factor 3 (basic) |
3848 |
0.17 |
| chr5_92236564_92236761 | 0.55 |
Ppef2 |
protein phosphatase, EF hand calcium-binding domain 2 |
7641 |
0.11 |
| chr11_96939648_96939799 | 0.55 |
Pnpo |
pyridoxine 5'-phosphate oxidase |
4056 |
0.1 |
| chr6_122391330_122392825 | 0.53 |
1700063H04Rik |
RIKEN cDNA 1700063H04 gene |
698 |
0.58 |
| chr1_160306832_160307632 | 0.53 |
Rabgap1l |
RAB GTPase activating protein 1-like |
107 |
0.95 |
| chr1_85933280_85934264 | 0.53 |
Gm28884 |
predicted gene 28884 |
3560 |
0.15 |
| chr1_173330607_173330937 | 0.53 |
Ackr1 |
atypical chemokine receptor 1 (Duffy blood group) |
2730 |
0.2 |
| chr2_38286593_38287645 | 0.52 |
Dennd1a |
DENN/MADD domain containing 1A |
69 |
0.96 |
| chr11_33202043_33204837 | 0.52 |
Tlx3 |
T cell leukemia, homeobox 3 |
149 |
0.86 |
| chr5_8894454_8894838 | 0.52 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
697 |
0.63 |
| chr7_67972762_67974418 | 0.52 |
Igf1r |
insulin-like growth factor I receptor |
20763 |
0.21 |
| chr9_66505109_66505930 | 0.52 |
Herc1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
5265 |
0.18 |
| chr11_46440435_46440727 | 0.52 |
Med7 |
mediator complex subunit 7 |
929 |
0.46 |
| chr15_102955712_102957246 | 0.52 |
Hoxc11 |
homeobox C11 |
2052 |
0.15 |
| chr17_33638155_33638934 | 0.51 |
Pram1 |
PML-RAR alpha-regulated adaptor molecule 1 |
449 |
0.7 |
| chr11_96199669_96200892 | 0.51 |
Hoxb13 |
homeobox B13 |
5964 |
0.11 |
| chr10_127177322_127177474 | 0.49 |
Slc26a10 |
solute carrier family 26, member 10 |
409 |
0.66 |
| chr4_41271157_41271308 | 0.49 |
Ubap2 |
ubiquitin-associated protein 2 |
3884 |
0.13 |
| chr17_36189137_36189654 | 0.48 |
H2-T3 |
histocompatibility 2, T region locus 3 |
727 |
0.28 |
| chr17_35194091_35194242 | 0.48 |
Ltb |
lymphotoxin B |
273 |
0.61 |
| chr17_25127118_25127513 | 0.47 |
Clcn7 |
chloride channel, voltage-sensitive 7 |
6076 |
0.1 |
| chr19_60871718_60871869 | 0.47 |
Prdx3 |
peroxiredoxin 3 |
2763 |
0.19 |
| chr17_28007198_28009699 | 0.46 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
1103 |
0.37 |
| chr14_25841885_25843222 | 0.46 |
Anxa11 |
annexin A11 |
370 |
0.81 |
| chr3_9767592_9767781 | 0.46 |
Gm16337 |
predicted gene 16337 |
10079 |
0.22 |
| chr16_68205625_68205776 | 0.46 |
Gm49650 |
predicted gene, 49650 |
40845 |
0.21 |
| chr19_4130763_4131308 | 0.45 |
Tmem134 |
transmembrane protein 134 |
3463 |
0.07 |
| chr6_91116004_91117353 | 0.45 |
Nup210 |
nucleoporin 210 |
118 |
0.95 |
| chr2_164432432_164433231 | 0.45 |
Sdc4 |
syndecan 4 |
10355 |
0.08 |
| chr5_97000494_97000645 | 0.44 |
Bmp2k |
BMP2 inducible kinase |
2880 |
0.18 |
| chr18_65802028_65802182 | 0.44 |
Sec11c |
SEC11 homolog C, signal peptidase complex subunit |
803 |
0.51 |
| chr9_62340666_62341173 | 0.43 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
374 |
0.87 |
| chr9_48362869_48363160 | 0.43 |
Nxpe4 |
neurexophilin and PC-esterase domain family, member 4 |
973 |
0.55 |
| chr3_51230467_51230944 | 0.43 |
Gm38357 |
predicted gene, 38357 |
1212 |
0.38 |
| chr4_135727528_135728972 | 0.43 |
Il22ra1 |
interleukin 22 receptor, alpha 1 |
78 |
0.96 |
| chr11_107992735_107992914 | 0.43 |
Gm11657 |
predicted gene 11657 |
1198 |
0.42 |
| chr15_78571734_78572724 | 0.42 |
Rac2 |
Rac family small GTPase 2 |
552 |
0.62 |
| chr7_16844794_16846016 | 0.42 |
Prkd2 |
protein kinase D2 |
172 |
0.9 |
| chr15_62158174_62158844 | 0.42 |
Pvt1 |
Pvt1 oncogene |
19666 |
0.25 |
| chr13_114342850_114343360 | 0.42 |
Ndufs4 |
NADH:ubiquinone oxidoreductase core subunit S4 |
44944 |
0.12 |
| chr4_132064709_132064928 | 0.42 |
Epb41 |
erythrocyte membrane protein band 4.1 |
7279 |
0.12 |
| chr7_28584162_28584331 | 0.42 |
Pak4 |
p21 (RAC1) activated kinase 4 |
13840 |
0.08 |
| chr5_143819014_143819653 | 0.42 |
Eif2ak1 |
eukaryotic translation initiation factor 2 alpha kinase 1 |
1386 |
0.38 |
| chr6_67161907_67162479 | 0.41 |
A430010J10Rik |
RIKEN cDNA A430010J10 gene |
2731 |
0.23 |
| chr6_87808529_87808706 | 0.41 |
Rab43 |
RAB43, member RAS oncogene family |
1137 |
0.24 |
| chr7_83651297_83652259 | 0.41 |
Il16 |
interleukin 16 |
3547 |
0.15 |
| chr2_39002991_39003196 | 0.41 |
Rpl35 |
ribosomal protein L35 |
1950 |
0.16 |
| chr10_118633408_118633559 | 0.41 |
Ifngas1 |
Ifng antisense RNA 1 |
76958 |
0.08 |
| chr16_93129255_93129653 | 0.41 |
Gm28003 |
predicted gene, 28003 |
56699 |
0.15 |
| chr15_50361344_50361976 | 0.40 |
Gm49198 |
predicted gene, 49198 |
74762 |
0.13 |
| chr4_141617213_141618510 | 0.40 |
Tmem82 |
transmembrane protein 82 |
120 |
0.93 |
| chr15_82166254_82166950 | 0.40 |
Srebf2 |
sterol regulatory element binding factor 2 |
18577 |
0.08 |
| chr5_139812274_139812441 | 0.39 |
Tmem184a |
transmembrane protein 184a |
886 |
0.46 |
| chr5_146686924_146687505 | 0.39 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
19408 |
0.16 |
| chr7_120879515_120880256 | 0.39 |
Eef2k |
eukaryotic elongation factor-2 kinase |
3665 |
0.15 |
| chr6_86078066_86079298 | 0.38 |
Add2 |
adducin 2 (beta) |
598 |
0.65 |
| chr5_145115755_145116764 | 0.38 |
Arpc1b |
actin related protein 2/3 complex, subunit 1B |
1943 |
0.18 |
| chr8_121082801_121085531 | 0.38 |
Foxf1 |
forkhead box F1 |
220 |
0.71 |
| chr13_117033409_117033738 | 0.38 |
Parp8 |
poly (ADP-ribose) polymerase family, member 8 |
8036 |
0.2 |
| chr5_105527486_105528761 | 0.38 |
Lrrc8c |
leucine rich repeat containing 8 family, member C |
8650 |
0.2 |
| chr18_32543718_32543992 | 0.38 |
Gypc |
glycophorin C |
8437 |
0.2 |
| chr9_50922410_50922924 | 0.37 |
Gm25558 |
predicted gene, 25558 |
34717 |
0.13 |
| chrX_143825863_143827628 | 0.37 |
Capn6 |
calpain 6 |
587 |
0.46 |
| chr18_65246605_65246956 | 0.37 |
Mir122 |
microRNA 122 |
2081 |
0.24 |
| chr9_46351879_46352155 | 0.37 |
Gm31374 |
predicted gene, 31374 |
9242 |
0.12 |
| chr11_96294128_96295421 | 0.36 |
Hoxb6 |
homeobox B6 |
2298 |
0.11 |
| chr1_136952214_136953356 | 0.36 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
845 |
0.67 |
| chr13_56748368_56748842 | 0.36 |
Gm45623 |
predicted gene 45623 |
5222 |
0.24 |
| chr1_191431994_191432741 | 0.36 |
Gm2272 |
predicted gene 2272 |
24701 |
0.12 |
| chr5_115531466_115532555 | 0.36 |
Pxn |
paxillin |
10904 |
0.09 |
| chr4_144423294_144423445 | 0.36 |
Pramef12 |
PRAME family member 12 |
14905 |
0.09 |
| chr15_76368582_76369737 | 0.36 |
Hgh1 |
HGH1 homolog |
261 |
0.77 |
| chr19_5724002_5724609 | 0.36 |
Ehbp1l1 |
EH domain binding protein 1-like 1 |
1965 |
0.11 |
| chrX_169915854_169917220 | 0.36 |
Mid1 |
midline 1 |
10414 |
0.22 |
| chr18_76242126_76242702 | 0.36 |
Smad2 |
SMAD family member 2 |
240 |
0.92 |
| chr2_36198146_36199404 | 0.36 |
Gm13429 |
predicted gene 13429 |
3539 |
0.16 |
| chr1_160049724_160049981 | 0.35 |
4930523C07Rik |
RIKEN cDNA 4930523C07 gene |
5405 |
0.17 |
| chr7_5029343_5032174 | 0.35 |
Zfp865 |
zinc finger protein 865 |
1434 |
0.15 |
| chr2_91635021_91635643 | 0.35 |
F2 |
coagulation factor II |
1063 |
0.36 |
| chr13_24947671_24947822 | 0.35 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
4594 |
0.15 |
| chr9_99874679_99877472 | 0.35 |
Sox14 |
SRY (sex determining region Y)-box 14 |
95 |
0.97 |
| chr3_9853920_9854071 | 0.35 |
Pag1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
20316 |
0.17 |
| chr10_94520068_94521446 | 0.35 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
5900 |
0.23 |
| chr7_101820668_101822582 | 0.35 |
Phox2a |
paired-like homeobox 2a |
879 |
0.38 |
| chr1_138579548_138580048 | 0.35 |
Gm37101 |
predicted gene, 37101 |
8442 |
0.18 |
| chr11_48873284_48874119 | 0.35 |
Irgm1 |
immunity-related GTPase family M member 1 |
2018 |
0.18 |
| chr2_122237098_122237979 | 0.34 |
Sord |
sorbitol dehydrogenase |
2789 |
0.15 |
| chr4_118544360_118544725 | 0.34 |
Tmem125 |
transmembrane protein 125 |
498 |
0.69 |
| chr2_74725203_74725767 | 0.34 |
Mir10b |
microRNA 10b |
585 |
0.37 |
| chr2_32061832_32061983 | 0.34 |
Gm16534 |
predicted gene 16534 |
2257 |
0.18 |
| chr11_102318146_102319152 | 0.34 |
Ubtf |
upstream binding transcription factor, RNA polymerase I |
447 |
0.7 |
| chr18_69665931_69667348 | 0.34 |
Tcf4 |
transcription factor 4 |
9972 |
0.29 |
| chr4_134868934_134869297 | 0.34 |
Rhd |
Rh blood group, D antigen |
4579 |
0.19 |
| chr16_18424546_18424973 | 0.34 |
Txnrd2 |
thioredoxin reductase 2 |
1625 |
0.21 |
| chr13_93388547_93389010 | 0.34 |
Gm47155 |
predicted gene, 47155 |
22005 |
0.14 |
| chr11_98581326_98581888 | 0.34 |
Ormdl3 |
ORM1-like 3 (S. cerevisiae) |
5761 |
0.11 |
| chr16_92489115_92489751 | 0.34 |
Clic6 |
chloride intracellular channel 6 |
3697 |
0.18 |
| chr6_103674240_103674525 | 0.34 |
Chl1 |
cell adhesion molecule L1-like |
23368 |
0.19 |
| chr12_117729655_117729806 | 0.34 |
Gm18955 |
predicted gene, 18955 |
5758 |
0.23 |
| chr10_82991418_82991936 | 0.33 |
Chst11 |
carbohydrate sulfotransferase 11 |
6179 |
0.19 |
| chr6_141091497_141091720 | 0.33 |
Gm7384 |
predicted gene 7384 |
46439 |
0.16 |
| chr11_40732046_40732503 | 0.33 |
Hmmr |
hyaluronan mediated motility receptor (RHAMM) |
1117 |
0.37 |
| chr2_167588684_167589456 | 0.33 |
Gm11475 |
predicted gene 11475 |
2325 |
0.19 |
| chr4_132040221_132040736 | 0.33 |
Gm13063 |
predicted gene 13063 |
7700 |
0.12 |
| chr7_118853034_118853185 | 0.33 |
Knop1 |
lysine rich nucleolar protein 1 |
2504 |
0.19 |
| chr9_72439224_72439615 | 0.33 |
Mns1 |
meiosis-specific nuclear structural protein 1 |
884 |
0.3 |
| chr16_49798750_49798901 | 0.33 |
Gm15518 |
predicted gene 15518 |
45 |
0.97 |
| chr17_33913300_33914237 | 0.33 |
Daxx |
Fas death domain-associated protein |
209 |
0.74 |
| chr4_108217222_108218259 | 0.33 |
Zyg11a |
zyg-11 family member A, cell cycle regulator |
182 |
0.94 |
| chr9_21015148_21016366 | 0.33 |
Icam1 |
intercellular adhesion molecule 1 |
228 |
0.81 |
| chr15_79209083_79209599 | 0.33 |
Gm10863 |
predicted gene 10863 |
30 |
0.95 |
| chr8_121570277_121571228 | 0.33 |
Fbxo31 |
F-box protein 31 |
7995 |
0.11 |
| chr11_48869797_48870455 | 0.32 |
Irgm1 |
immunity-related GTPase family M member 1 |
1248 |
0.29 |
| chr9_44340460_44342952 | 0.32 |
Hmbs |
hydroxymethylbilane synthase |
473 |
0.51 |
| chr15_81585550_81586926 | 0.32 |
Ep300 |
E1A binding protein p300 |
18 |
0.58 |
| chr15_91610712_91610863 | 0.32 |
4933438A12Rik |
RIKEN cDNA 4933438A12 gene |
13204 |
0.17 |
| chr3_144759210_144759685 | 0.32 |
Clca3a1 |
chloride channel accessory 3A1 |
1394 |
0.3 |
| chr16_92699485_92700446 | 0.32 |
Runx1 |
runt related transcription factor 1 |
2637 |
0.36 |
| chr5_64803350_64804590 | 0.32 |
C230096K16Rik |
RIKEN cDNA C230096K16 gene |
44 |
0.56 |
| chr12_108855592_108855779 | 0.32 |
Slc25a47 |
solute carrier family 25, member 47 |
4546 |
0.11 |
| chr3_145987511_145989209 | 0.31 |
Syde2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
223 |
0.93 |
| chr4_3877971_3878122 | 0.31 |
Mos |
Moloney sarcoma oncogene |
5941 |
0.14 |
| chr1_155736856_155738497 | 0.31 |
Lhx4 |
LIM homeobox protein 4 |
4351 |
0.18 |
| chr8_61731359_61731510 | 0.31 |
Palld |
palladin, cytoskeletal associated protein |
28655 |
0.22 |
| chr8_122480707_122480858 | 0.31 |
Ctu2 |
cytosolic thiouridylase subunit 2 |
140 |
0.89 |
| chr14_44033999_44034275 | 0.31 |
Gm3287 |
predicted gene 3287 |
586 |
0.5 |
| chr1_89921735_89923880 | 0.31 |
Gbx2 |
gastrulation brain homeobox 2 |
6378 |
0.21 |
| chr4_49481203_49481354 | 0.31 |
Gm26424 |
predicted gene, 26424 |
6971 |
0.13 |
| chr14_67263564_67263804 | 0.31 |
Gm23178 |
predicted gene, 23178 |
11279 |
0.16 |
| chr9_63144078_63146888 | 0.31 |
Skor1 |
SKI family transcriptional corepressor 1 |
1497 |
0.38 |
| chr16_23061643_23061794 | 0.31 |
Kng1 |
kininogen 1 |
3325 |
0.1 |
| chr7_67655369_67656000 | 0.31 |
Ttc23 |
tetratricopeptide repeat domain 23 |
237 |
0.9 |
| chr4_57914999_57916744 | 0.31 |
D630039A03Rik |
RIKEN cDNA D630039A03 gene |
426 |
0.84 |
| chr16_92058346_92059208 | 0.31 |
Mrps6 |
mitochondrial ribosomal protein S6 |
36 |
0.8 |
| chr13_31546831_31546982 | 0.31 |
Foxq1 |
forkhead box Q1 |
9228 |
0.14 |
| chr8_121791189_121791340 | 0.30 |
Klhdc4 |
kelch domain containing 4 |
6164 |
0.13 |
| chr5_130016232_130016383 | 0.30 |
Asl |
argininosuccinate lyase |
1585 |
0.26 |
| chr10_81563960_81565660 | 0.30 |
Tle5 |
TLE family member 5, transcriptional modulator |
205 |
0.84 |
| chr2_162967542_162967693 | 0.30 |
Gm11453 |
predicted gene 11453 |
15644 |
0.11 |
| chr1_153413763_153413914 | 0.30 |
E330020D12Rik |
Riken cDNA E330020D12 gene |
395 |
0.81 |
| chr6_52239036_52239383 | 0.30 |
Hoxa10 |
homeobox A10 |
1586 |
0.13 |
| chr4_77911888_77912039 | 0.30 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
299776 |
0.01 |
| chr7_118742325_118742557 | 0.30 |
1700016B15Rik |
RIKEN cDNA 1700016B15 gene |
1260 |
0.29 |
| chr7_68189508_68190188 | 0.30 |
Igf1r |
insulin-like growth factor I receptor |
3223 |
0.28 |
| chr10_59873887_59874700 | 0.30 |
Gm7413 |
predicted gene 7413 |
2611 |
0.21 |
| chr17_26117534_26117685 | 0.29 |
Tmem8 |
transmembrane protein 8 |
220 |
0.84 |
| chr19_9954351_9955257 | 0.29 |
Gm50346 |
predicted gene, 50346 |
11972 |
0.09 |
| chr4_106924547_106925049 | 0.29 |
Ssbp3 |
single-stranded DNA binding protein 3 |
1573 |
0.42 |
| chr16_44174924_44175152 | 0.29 |
Usf3 |
upstream transcription factor family member 3 |
1641 |
0.38 |
| chr14_30942231_30942467 | 0.29 |
Itih1 |
inter-alpha trypsin inhibitor, heavy chain 1 |
928 |
0.4 |
| chr19_34744341_34744492 | 0.29 |
Slc16a12 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
2873 |
0.17 |
| chrX_101256637_101256923 | 0.29 |
Foxo4 |
forkhead box O4 |
1959 |
0.16 |
| chr9_107375414_107375565 | 0.29 |
4930429P21Rik |
RIKEN cDNA 4930429P21 gene |
21503 |
0.09 |
| chr4_44778425_44778576 | 0.29 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
18056 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.4 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.1 | 0.9 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.4 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.1 | 0.2 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.2 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.4 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.1 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.4 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.2 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.5 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.2 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.5 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.0 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) |
| 0.0 | 0.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.0 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.0 | GO:0048819 | regulation of hair follicle maturation(GO:0048819) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.0 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.0 | 0.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060717 | chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0051446 | positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0014870 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.0 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0034805 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 1.8 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.0 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |