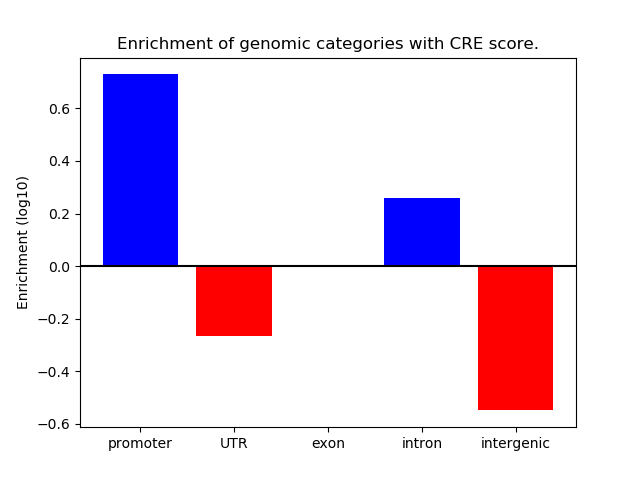
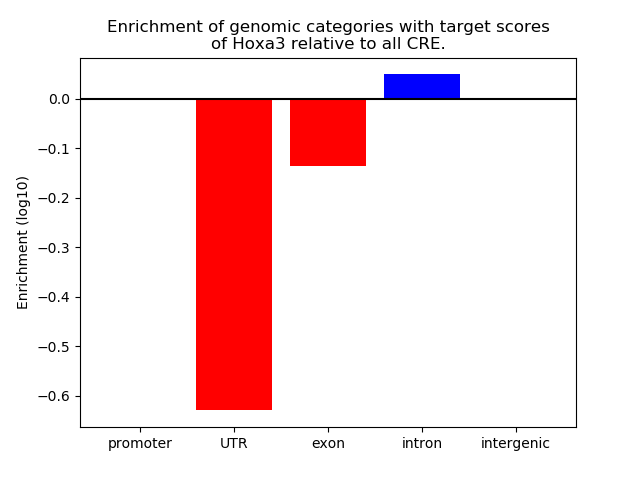
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
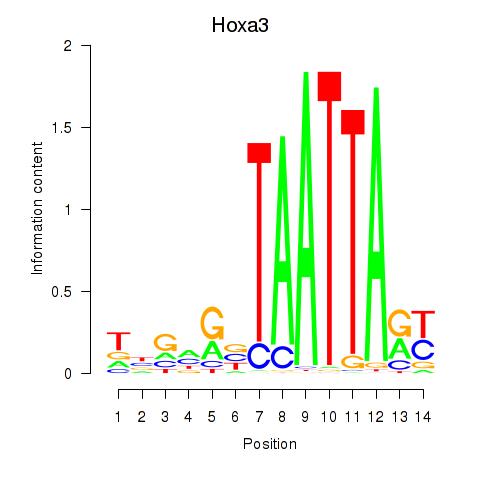
Results for Hoxa3
Z-value: 0.36
Transcription factors associated with Hoxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa3
|
ENSMUSG00000079560.7 | homeobox A3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_52211753_52213405 | Hoxa3 | 536 | 0.359745 | 0.14 | 3.0e-01 | Click! |
| chr6_52185807_52185988 | Hoxa3 | 1023 | 0.205032 | -0.12 | 3.6e-01 | Click! |
| chr6_52187599_52188482 | Hoxa3 | 3166 | 0.068417 | 0.09 | 5.1e-01 | Click! |
| chr6_52211085_52211739 | Hoxa3 | 1703 | 0.105183 | 0.08 | 5.4e-01 | Click! |
| chr6_52187293_52187465 | Hoxa3 | 2505 | 0.080085 | -0.08 | 5.7e-01 | Click! |
Activity of the Hoxa3 motif across conditions
Conditions sorted by the z-value of the Hoxa3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_33083500_33083959 | 0.94 |
Trdn |
triadin |
168 |
0.97 |
| chr6_146500226_146500471 | 0.67 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
1530 |
0.4 |
| chr8_57317953_57318771 | 0.58 |
Hand2os1 |
Hand2, opposite strand 1 |
462 |
0.73 |
| chr2_20942943_20943436 | 0.54 |
Arhgap21 |
Rho GTPase activating protein 21 |
230 |
0.93 |
| chr12_117200442_117200898 | 0.51 |
Gm10421 |
predicted gene 10421 |
49019 |
0.16 |
| chrX_142684293_142684706 | 0.47 |
Tmem164 |
transmembrane protein 164 |
1611 |
0.4 |
| chr14_63249784_63250324 | 0.46 |
Gata4 |
GATA binding protein 4 |
4783 |
0.2 |
| chr5_148990792_148991310 | 0.45 |
Gm42791 |
predicted gene 42791 |
4096 |
0.1 |
| chr13_44167401_44167573 | 0.44 |
Gm33684 |
predicted gene, 33684 |
16892 |
0.17 |
| chr5_116456914_116457389 | 0.42 |
Gm42853 |
predicted gene 42853 |
333 |
0.83 |
| chr10_45338438_45338763 | 0.41 |
Bves |
blood vessel epicardial substance |
2828 |
0.24 |
| chr2_79087179_79088090 | 0.39 |
Gm14469 |
predicted gene 14469 |
48900 |
0.14 |
| chr8_61901693_61901844 | 0.37 |
Palld |
palladin, cytoskeletal associated protein |
901 |
0.56 |
| chr5_138609015_138609166 | 0.37 |
Gm15497 |
predicted gene 15497 |
1649 |
0.2 |
| chr11_65269494_65270784 | 0.37 |
Myocd |
myocardin |
150 |
0.96 |
| chr5_143293458_143294037 | 0.37 |
Zfp853 |
zinc finger protein 853 |
124 |
0.91 |
| chr11_65266805_65267363 | 0.36 |
Myocd |
myocardin |
2770 |
0.29 |
| chr3_155052033_155052416 | 0.36 |
Tnni3k |
TNNI3 interacting kinase |
3126 |
0.26 |
| chr13_46424530_46424922 | 0.36 |
Rbm24 |
RNA binding motif protein 24 |
2904 |
0.31 |
| chr7_29819755_29819906 | 0.35 |
Zfp790 |
zinc finger protein 790 |
839 |
0.4 |
| chr12_51594696_51594847 | 0.35 |
Coch |
cochlin |
1392 |
0.48 |
| chr10_39165752_39166209 | 0.34 |
Ccn6 |
cellular communication network factor 6 |
2186 |
0.23 |
| chr10_108944251_108944402 | 0.33 |
Gm47477 |
predicted gene, 47477 |
12162 |
0.27 |
| chr15_81873813_81874521 | 0.33 |
Aco2 |
aconitase 2, mitochondrial |
1477 |
0.21 |
| chrX_138915472_138915880 | 0.33 |
Nrk |
Nik related kinase |
1245 |
0.61 |
| chr4_87359673_87359861 | 0.32 |
Gm12600 |
predicted gene 12600 |
61405 |
0.13 |
| chr2_63982565_63982778 | 0.32 |
Fign |
fidgetin |
115317 |
0.07 |
| chr13_41486298_41486655 | 0.31 |
Gm32401 |
predicted gene, 32401 |
654 |
0.45 |
| chr2_48417232_48417383 | 0.31 |
Gm13472 |
predicted gene 13472 |
29270 |
0.17 |
| chrX_85775362_85775513 | 0.30 |
Gk |
glycerol kinase |
1187 |
0.39 |
| chrX_146946268_146946419 | 0.30 |
Gm4996 |
predicted gene 4996 |
13658 |
0.15 |
| chr18_25694097_25694278 | 0.30 |
0710001A04Rik |
RIKEN cDNA 0710001A04 gene |
19583 |
0.23 |
| chr10_95297621_95297998 | 0.30 |
Gm48880 |
predicted gene, 48880 |
17044 |
0.14 |
| chrX_53267645_53268215 | 0.30 |
Fam122b |
family with sequence similarity 122, member B |
1090 |
0.45 |
| chr8_30958732_30959710 | 0.30 |
Gm45252 |
predicted gene 45252 |
44687 |
0.18 |
| chr16_44015370_44016774 | 0.28 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr7_44472607_44472821 | 0.28 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
824 |
0.31 |
| chr14_32684282_32684433 | 0.28 |
3425401B19Rik |
RIKEN cDNA 3425401B19 gene |
915 |
0.46 |
| chr2_172937060_172938226 | 0.28 |
Bmp7 |
bone morphogenetic protein 7 |
2449 |
0.28 |
| chr18_4636274_4636995 | 0.28 |
Jcad |
junctional cadherin 5 associated |
1756 |
0.43 |
| chr1_177199086_177199237 | 0.28 |
Gm37706 |
predicted gene, 37706 |
28299 |
0.14 |
| chr5_24350563_24350755 | 0.27 |
Kcnh2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
945 |
0.38 |
| chr1_46970388_46970697 | 0.26 |
Gm28527 |
predicted gene 28527 |
73663 |
0.09 |
| chr14_65374778_65375936 | 0.26 |
Zfp395 |
zinc finger protein 395 |
36 |
0.97 |
| chr5_27264978_27265420 | 0.25 |
Dpp6 |
dipeptidylpeptidase 6 |
3224 |
0.31 |
| chr14_46491764_46491915 | 0.25 |
Gm34756 |
predicted gene, 34756 |
10611 |
0.12 |
| chr1_140182332_140182724 | 0.25 |
Cfh |
complement component factor h |
752 |
0.73 |
| chr6_117605384_117605669 | 0.25 |
Gm45083 |
predicted gene 45083 |
7044 |
0.22 |
| chr12_112590746_112590946 | 0.24 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
1580 |
0.31 |
| chr2_152436557_152436921 | 0.24 |
6820408C15Rik |
RIKEN cDNA 6820408C15 gene |
9077 |
0.08 |
| chr10_72299004_72299155 | 0.24 |
Gm9923 |
predicted pseudogene 9923 |
10242 |
0.31 |
| chr1_186515830_186515981 | 0.24 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
42567 |
0.18 |
| chr16_96205810_96206739 | 0.24 |
Sh3bgr |
SH3-binding domain glutamic acid-rich protein |
369 |
0.83 |
| chr15_40661942_40662224 | 0.24 |
Zfpm2 |
zinc finger protein, multitype 2 |
2907 |
0.38 |
| chr9_86438288_86439112 | 0.23 |
Ube2cbp |
ubiquitin-conjugating enzyme E2C binding protein |
26250 |
0.17 |
| chr1_89991921_89993000 | 0.23 |
Asb18 |
ankyrin repeat and SOCS box-containing 18 |
3992 |
0.25 |
| chr13_45571916_45572112 | 0.23 |
Gmpr |
guanosine monophosphate reductase |
25861 |
0.23 |
| chr11_117482269_117483090 | 0.22 |
Gm34418 |
predicted gene, 34418 |
82 |
0.95 |
| chr4_105100875_105101089 | 0.22 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
8908 |
0.27 |
| chr4_32486662_32487238 | 0.22 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
14555 |
0.23 |
| chr16_72699332_72699617 | 0.22 |
Robo1 |
roundabout guidance receptor 1 |
36270 |
0.24 |
| chr8_109417937_109418404 | 0.22 |
Gm23163 |
predicted gene, 23163 |
7574 |
0.24 |
| chr9_77919114_77919265 | 0.22 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
1755 |
0.29 |
| chr10_33091664_33091948 | 0.22 |
Trdn |
triadin |
8245 |
0.26 |
| chr5_52806998_52807149 | 0.21 |
Zcchc4 |
zinc finger, CCHC domain containing 4 |
2747 |
0.27 |
| chr14_46202990_46203141 | 0.21 |
Ubb-ps |
ubiquitin B, pseudogene |
118937 |
0.05 |
| chr5_138608490_138608879 | 0.21 |
Zfp68 |
zinc finger protein 68 |
1627 |
0.2 |
| chr10_60826340_60827679 | 0.21 |
Unc5b |
unc-5 netrin receptor B |
4373 |
0.21 |
| chr7_136007847_136008710 | 0.21 |
Gm9341 |
predicted gene 9341 |
55480 |
0.13 |
| chr2_94404907_94405280 | 0.21 |
Ttc17 |
tetratricopeptide repeat domain 17 |
102 |
0.94 |
| chr13_71961770_71962476 | 0.21 |
Irx1 |
Iroquois homeobox 1 |
1407 |
0.5 |
| chr9_79976568_79977294 | 0.21 |
Filip1 |
filamin A interacting protein 1 |
728 |
0.66 |
| chr9_47650354_47650592 | 0.21 |
Cadm1 |
cell adhesion molecule 1 |
120100 |
0.05 |
| chr8_85427729_85428024 | 0.20 |
1700051O22Rik |
RIKEN cDNA 1700051O22 Gene |
4810 |
0.15 |
| chr19_44631481_44631730 | 0.20 |
Gm35460 |
predicted gene, 35460 |
16840 |
0.15 |
| chr1_77708269_77708811 | 0.20 |
Gm28385 |
predicted gene 28385 |
137868 |
0.05 |
| chr14_27237814_27239092 | 0.20 |
Gm49616 |
predicted gene, 49616 |
391 |
0.49 |
| chr11_52166541_52167570 | 0.20 |
Olfr1372-ps1 |
olfactory receptor 1372, pseudogene 1 |
1104 |
0.37 |
| chr4_116073957_116074118 | 0.19 |
Uqcrh |
ubiquinol-cytochrome c reductase hinge protein |
994 |
0.31 |
| chr4_5724213_5725550 | 0.19 |
Fam110b |
family with sequence similarity 110, member B |
569 |
0.81 |
| chr18_56683771_56684023 | 0.19 |
Gm50289 |
predicted gene, 50289 |
16119 |
0.16 |
| chr7_136275874_136276030 | 0.19 |
C030029H02Rik |
RIKEN cDNA C030029H02 gene |
7618 |
0.21 |
| chr6_88041235_88041766 | 0.19 |
Gm44187 |
predicted gene, 44187 |
967 |
0.4 |
| chr16_4448299_4448450 | 0.19 |
Adcy9 |
adenylate cyclase 9 |
27876 |
0.16 |
| chr15_11741805_11742688 | 0.19 |
Gm41271 |
predicted gene, 41271 |
38543 |
0.18 |
| chr10_45048086_45048775 | 0.18 |
Prep |
prolyl endopeptidase |
18773 |
0.15 |
| chr11_88617275_88617426 | 0.18 |
Msi2 |
musashi RNA-binding protein 2 |
27203 |
0.2 |
| chr14_79645123_79645681 | 0.18 |
Cnmd |
chondromodulin |
108 |
0.96 |
| chr1_72226384_72226568 | 0.18 |
Gm25360 |
predicted gene, 25360 |
236 |
0.89 |
| chr5_137348685_137349851 | 0.18 |
Ephb4 |
Eph receptor B4 |
841 |
0.39 |
| chr3_8684903_8685755 | 0.18 |
Gm23670 |
predicted gene, 23670 |
5852 |
0.16 |
| chr9_98964920_98965344 | 0.18 |
Foxl2os |
forkhead box L2, opposite strand |
9833 |
0.12 |
| chr14_54362334_54362485 | 0.17 |
Oxa1l |
oxidase assembly 1-like |
1364 |
0.25 |
| chr10_117044417_117044727 | 0.17 |
Lrrc10 |
leucine rich repeat containing 10 |
769 |
0.38 |
| chr17_71290536_71290920 | 0.17 |
Emilin2 |
elastin microfibril interfacer 2 |
6519 |
0.15 |
| chr8_90176316_90176467 | 0.17 |
Tox3 |
TOX high mobility group box family member 3 |
171735 |
0.04 |
| chr11_117505657_117505808 | 0.17 |
Gm11734 |
predicted gene 11734 |
17871 |
0.14 |
| chr8_92361507_92362918 | 0.17 |
Irx5 |
Iroquois homeobox 5 |
4463 |
0.2 |
| chr3_148983311_148983462 | 0.17 |
Gm43573 |
predicted gene 43573 |
5766 |
0.19 |
| chr3_126599850_126600378 | 0.17 |
Gm43011 |
predicted gene 43011 |
85 |
0.96 |
| chr8_80853437_80854023 | 0.16 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
22846 |
0.16 |
| chr3_65832584_65832735 | 0.16 |
Gm37131 |
predicted gene, 37131 |
32170 |
0.13 |
| chr9_103364095_103364246 | 0.16 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
727 |
0.6 |
| chr8_27263249_27263470 | 0.16 |
Eif4ebp1 |
eukaryotic translation initiation factor 4E binding protein 1 |
1055 |
0.37 |
| chr3_66976156_66976605 | 0.16 |
Shox2 |
short stature homeobox 2 |
2067 |
0.29 |
| chr13_110396048_110396669 | 0.16 |
Plk2 |
polo like kinase 2 |
346 |
0.91 |
| chr1_32176216_32176451 | 0.16 |
Khdrbs2 |
KH domain containing, RNA binding, signal transduction associated 2 |
3446 |
0.32 |
| chr13_89547281_89547432 | 0.16 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
7560 |
0.25 |
| chr10_120128769_120129086 | 0.16 |
Helb |
helicase (DNA) B |
15940 |
0.17 |
| chr2_64092860_64093848 | 0.16 |
Fign |
fidgetin |
4634 |
0.37 |
| chr7_50588908_50589191 | 0.16 |
4933405O20Rik |
RIKEN cDNA 4933405O20 gene |
10141 |
0.24 |
| chr6_144419028_144419519 | 0.16 |
Sox5os2 |
SRY (sex determining region Y)-box 5, opposite strand 2 |
61952 |
0.15 |
| chr14_119831360_119832142 | 0.16 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
32773 |
0.18 |
| chr14_119602272_119602445 | 0.15 |
Gm6212 |
predicted gene 6212 |
41654 |
0.2 |
| chr19_45223479_45224523 | 0.15 |
Lbx1 |
ladybird homeobox 1 |
11811 |
0.18 |
| chr13_9494460_9495468 | 0.15 |
Gm48871 |
predicted gene, 48871 |
49064 |
0.12 |
| chr3_70104094_70104909 | 0.15 |
Gm37585 |
predicted gene, 37585 |
12031 |
0.22 |
| chr3_87630903_87631116 | 0.15 |
Arhgef11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
12248 |
0.16 |
| chr19_45235899_45236901 | 0.15 |
Lbx1 |
ladybird homeobox 1 |
588 |
0.75 |
| chr2_101652616_101653044 | 0.15 |
B230118H07Rik |
RIKEN cDNA B230118H07 gene |
3298 |
0.2 |
| chr9_28056769_28056920 | 0.15 |
Gm15606 |
predicted gene 15606 |
141043 |
0.05 |
| chr3_55782023_55782190 | 0.15 |
Mab21l1 |
mab-21-like 1 |
404 |
0.8 |
| chr6_45096373_45096524 | 0.15 |
Gm43879 |
predicted gene, 43879 |
15702 |
0.16 |
| chr5_103972357_103973054 | 0.15 |
Hsd17b13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
4621 |
0.16 |
| chr14_20696186_20696337 | 0.15 |
Fut11 |
fucosyltransferase 11 |
1284 |
0.22 |
| chr6_97457802_97459465 | 0.15 |
Frmd4b |
FERM domain containing 4B |
653 |
0.74 |
| chr13_115098735_115099036 | 0.15 |
Gm49395 |
predicted gene, 49395 |
2739 |
0.18 |
| chr4_44083540_44083707 | 0.15 |
Gne |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
300 |
0.84 |
| chr10_37988715_37989307 | 0.14 |
Gm24710 |
predicted gene, 24710 |
57331 |
0.15 |
| chr16_4519120_4519560 | 0.14 |
Srl |
sarcalumenin |
3723 |
0.19 |
| chr9_65162369_65163541 | 0.14 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
17783 |
0.12 |
| chr19_29271229_29271592 | 0.14 |
Jak2 |
Janus kinase 2 |
18969 |
0.16 |
| chr5_105052746_105052954 | 0.14 |
Gbp8 |
guanylate-binding protein 8 |
706 |
0.66 |
| chr1_136868933_136869351 | 0.14 |
Gm17781 |
predicted gene, 17781 |
70183 |
0.08 |
| chr8_119365249_119365400 | 0.14 |
Gm45723 |
predicted gene 45723 |
13372 |
0.16 |
| chr8_34079766_34081014 | 0.14 |
Dctn6 |
dynactin 6 |
12535 |
0.12 |
| chr8_57799694_57800553 | 0.14 |
Galntl6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
3533 |
0.27 |
| chr10_121504972_121505664 | 0.14 |
Gm46204 |
predicted gene, 46204 |
33 |
0.95 |
| chr18_35844890_35845340 | 0.14 |
Cxxc5 |
CXXC finger 5 |
9572 |
0.1 |
| chr6_91980058_91980209 | 0.14 |
Fgd5 |
FYVE, RhoGEF and PH domain containing 5 |
1255 |
0.39 |
| chr11_19012477_19012923 | 0.14 |
Meis1 |
Meis homeobox 1 |
698 |
0.49 |
| chr15_50870065_50870337 | 0.14 |
Trps1 |
transcriptional repressor GATA binding 1 |
12605 |
0.21 |
| chr1_161494611_161494762 | 0.13 |
Tnfsf18 |
tumor necrosis factor (ligand) superfamily, member 18 |
31 |
0.98 |
| chr1_81593421_81594539 | 0.13 |
Gm6198 |
predicted gene 6198 |
36497 |
0.2 |
| chr12_9253584_9253735 | 0.13 |
Gm46323 |
predicted gene, 46323 |
117779 |
0.06 |
| chr2_172448714_172448865 | 0.13 |
Rtf2 |
replication termination factor 2 |
4188 |
0.15 |
| chr13_30724955_30725406 | 0.13 |
Gm11370 |
predicted gene 11370 |
6597 |
0.21 |
| chr3_63899898_63900049 | 0.13 |
Plch1 |
phospholipase C, eta 1 |
501 |
0.73 |
| chr9_79977295_79978179 | 0.13 |
Filip1 |
filamin A interacting protein 1 |
67 |
0.97 |
| chr13_73262153_73264451 | 0.13 |
Irx4 |
Iroquois homeobox 4 |
2805 |
0.22 |
| chr11_64656679_64656830 | 0.13 |
Gm24275 |
predicted gene, 24275 |
66114 |
0.13 |
| chr12_107010953_107011720 | 0.13 |
Gm16087 |
predicted gene 16087 |
55408 |
0.13 |
| chr1_155526845_155527426 | 0.13 |
Gm5532 |
predicted gene 5532 |
8 |
0.98 |
| chr6_148598605_148599513 | 0.13 |
Gm6313 |
predicted gene 6313 |
10674 |
0.2 |
| chr8_34890130_34891317 | 0.13 |
Tnks |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
572 |
0.8 |
| chr4_147131860_147132011 | 0.13 |
Zfp991 |
zinc finger protein 991 |
103 |
0.94 |
| chr13_109262801_109262952 | 0.13 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
2222 |
0.44 |
| chr8_109250884_109251908 | 0.13 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
1530 |
0.52 |
| chr6_98525524_98525718 | 0.12 |
4930595L18Rik |
RIKEN cDNA 4930595L18 gene |
26486 |
0.26 |
| chr11_76334808_76334959 | 0.12 |
Nxn |
nucleoredoxin |
64185 |
0.09 |
| chr2_145233751_145234233 | 0.12 |
Slc24a3 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
8619 |
0.29 |
| chr2_167713707_167713858 | 0.12 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
22601 |
0.11 |
| chr8_94152224_94153713 | 0.12 |
Mt3 |
metallothionein 3 |
222 |
0.87 |
| chr10_69690251_69691139 | 0.12 |
Ank3 |
ankyrin 3, epithelial |
15601 |
0.28 |
| chr9_60742965_60743393 | 0.12 |
1700036A12Rik |
RIKEN cDNA 1700036A12 gene |
59 |
0.97 |
| chr12_98577628_98578516 | 0.12 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
238 |
0.9 |
| chr11_96341535_96342715 | 0.12 |
Gm11536 |
predicted gene 11536 |
17 |
0.92 |
| chr2_84455336_84455529 | 0.12 |
Tfpi |
tissue factor pathway inhibitor |
2678 |
0.26 |
| chr3_88509652_88510583 | 0.12 |
Lmna |
lamin A |
161 |
0.88 |
| chr8_61895218_61895438 | 0.12 |
Palld |
palladin, cytoskeletal associated protein |
7341 |
0.18 |
| chr4_40851422_40852119 | 0.12 |
Gm25931 |
predicted gene, 25931 |
1368 |
0.21 |
| chr10_68191368_68192460 | 0.12 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
55288 |
0.14 |
| chr19_45228695_45230660 | 0.12 |
Lbx1 |
ladybird homeobox 1 |
6135 |
0.2 |
| chr3_155054773_155054924 | 0.12 |
Tnni3k |
TNNI3 interacting kinase |
502 |
0.8 |
| chr6_50358058_50358744 | 0.12 |
Osbpl3 |
oxysterol binding protein-like 3 |
6444 |
0.27 |
| chr11_75291523_75292140 | 0.12 |
Gm47300 |
predicted gene, 47300 |
26777 |
0.12 |
| chr8_110321934_110322150 | 0.12 |
Gm26832 |
predicted gene, 26832 |
54580 |
0.11 |
| chr8_48596222_48596652 | 0.11 |
Gm45772 |
predicted gene 45772 |
34732 |
0.19 |
| chr19_34251985_34252204 | 0.11 |
Acta2 |
actin, alpha 2, smooth muscle, aorta |
2136 |
0.26 |
| chr13_12033129_12034063 | 0.11 |
Gm47493 |
predicted gene, 47493 |
18459 |
0.21 |
| chr4_145585026_145585604 | 0.11 |
Gm13212 |
predicted gene 13212 |
112 |
0.93 |
| chr18_12741375_12742056 | 0.11 |
Cabyr |
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
360 |
0.78 |
| chr5_150261018_150262108 | 0.11 |
Fry |
FRY microtubule binding protein |
1796 |
0.34 |
| chrX_106921477_106921682 | 0.11 |
Lpar4 |
lysophosphatidic acid receptor 4 |
954 |
0.64 |
| chr17_66276732_66277218 | 0.11 |
Gm49932 |
predicted gene, 49932 |
40606 |
0.1 |
| chr12_60595918_60596185 | 0.11 |
Gm6068 |
predicted gene 6068 |
47813 |
0.16 |
| chr16_49408257_49408911 | 0.11 |
Gm4802 |
predicted gene 4802 |
4046 |
0.24 |
| chr14_56402597_56403081 | 0.11 |
Rnf17 |
ring finger protein 17 |
139 |
0.94 |
| chr5_105878641_105878792 | 0.11 |
Zfp326 |
zinc finger protein 326 |
54 |
0.93 |
| chr16_72807953_72808738 | 0.11 |
Robo1 |
roundabout guidance receptor 1 |
145141 |
0.05 |
| chr16_25945207_25945588 | 0.11 |
Gm4524 |
predicted gene 4524 |
11078 |
0.24 |
| chr4_84637359_84638298 | 0.11 |
Bnc2 |
basonuclin 2 |
37168 |
0.19 |
| chr10_67056195_67056966 | 0.11 |
Reep3 |
receptor accessory protein 3 |
19258 |
0.17 |
| chr14_66325170_66325451 | 0.11 |
Gm41183 |
predicted gene, 41183 |
15919 |
0.14 |
| chr11_108336593_108336967 | 0.11 |
Apoh |
apolipoprotein H |
6574 |
0.19 |
| chr12_9570099_9571217 | 0.11 |
1700022H16Rik |
RIKEN cDNA 1700022H16 gene |
73 |
0.9 |
| chr2_19014190_19014343 | 0.11 |
4930426L09Rik |
RIKEN cDNA 4930426L09 gene |
15923 |
0.2 |
| chrX_153037586_153038140 | 0.11 |
9530051G07Rik |
RIKEN cDNA 9530051G07 gene |
234 |
0.55 |
| chr9_57158351_57158614 | 0.11 |
Commd4 |
COMM domain containing 4 |
151 |
0.93 |
| chr13_67332918_67333177 | 0.11 |
Zfp595 |
zinc finger protein 595 |
477 |
0.61 |
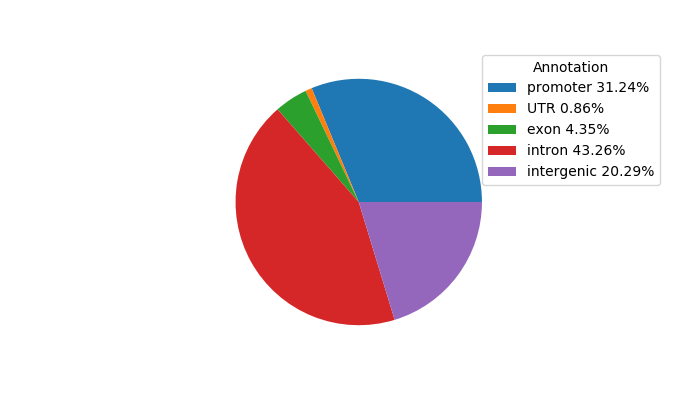
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.5 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.2 | GO:0003195 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.0 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |