Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
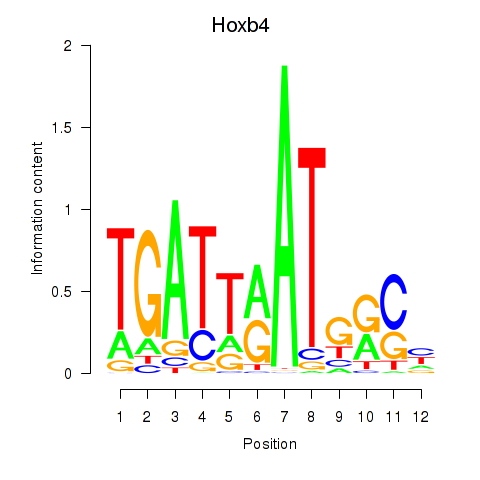
Results for Hoxb4
Z-value: 0.29
Transcription factors associated with Hoxb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb4
|
ENSMUSG00000038692.7 | homeobox B4 |
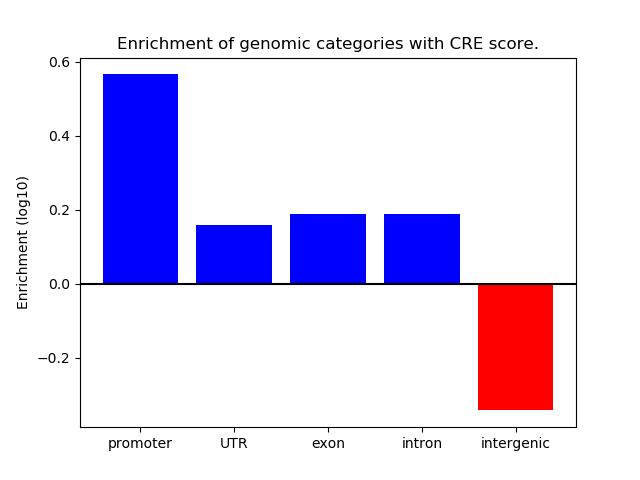
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_96318060_96319501 | Hoxb4 | 513 | 0.505895 | 0.06 | 6.5e-01 | Click! |
| chr11_96319562_96320434 | Hoxb4 | 1731 | 0.139807 | -0.01 | 9.2e-01 | Click! |
Activity of the Hoxb4 motif across conditions
Conditions sorted by the z-value of the Hoxb4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
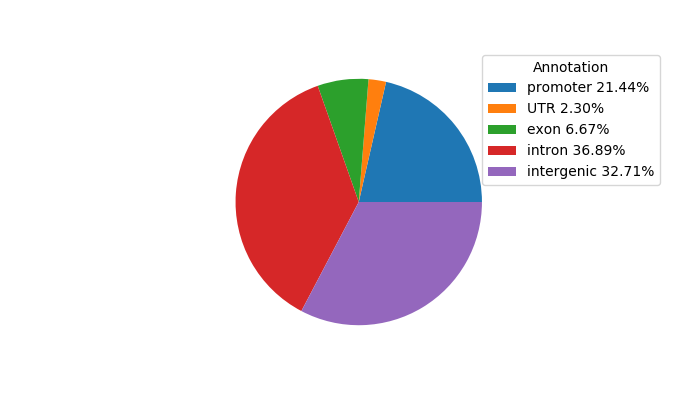
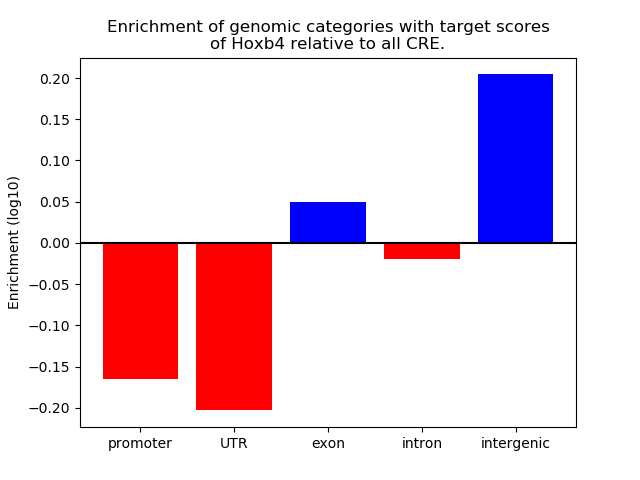
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_168426195_168428871 | 0.29 |
Pbx1 |
pre B cell leukemia homeobox 1 |
3971 |
0.3 |
| chr11_117454277_117454520 | 0.27 |
Gm34418 |
predicted gene, 34418 |
28199 |
0.11 |
| chr3_35332632_35333522 | 0.26 |
Gm25442 |
predicted gene, 25442 |
7463 |
0.26 |
| chr1_190017944_190018756 | 0.26 |
Smyd2 |
SET and MYND domain containing 2 |
95987 |
0.07 |
| chr10_102513522_102514467 | 0.25 |
Rassf9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
1738 |
0.36 |
| chr11_117600891_117601042 | 0.22 |
2900041M22Rik |
RIKEN cDNA 2900041M22 gene |
10281 |
0.19 |
| chr5_35466165_35466783 | 0.21 |
Cpz |
carboxypeptidase Z |
59097 |
0.08 |
| chr11_24740194_24740345 | 0.19 |
Gm10466 |
predicted gene 10466 |
9547 |
0.27 |
| chr2_140668225_140669392 | 0.19 |
Flrt3 |
fibronectin leucine rich transmembrane protein 3 |
2592 |
0.43 |
| chr3_30009457_30010152 | 0.19 |
Mecom |
MDS1 and EVI1 complex locus |
1635 |
0.36 |
| chr1_72038129_72038714 | 0.18 |
4930556G22Rik |
RIKEN cDNA 4930556G22 gene |
310 |
0.88 |
| chr11_96367227_96367378 | 0.18 |
Hoxb1 |
homeobox B1 |
1044 |
0.31 |
| chr2_172934875_172936042 | 0.17 |
Bmp7 |
bone morphogenetic protein 7 |
4634 |
0.21 |
| chr9_57803673_57803954 | 0.17 |
Arid3b |
AT rich interactive domain 3B (BRIGHT-like) |
7592 |
0.16 |
| chr18_4137682_4137958 | 0.16 |
Lyzl1 |
lysozyme-like 1 |
28012 |
0.19 |
| chr17_43360343_43362046 | 0.16 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
743 |
0.75 |
| chr17_84082064_84082337 | 0.16 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
3540 |
0.19 |
| chr13_15468099_15468909 | 0.16 |
Gli3 |
GLI-Kruppel family member GLI3 |
4524 |
0.2 |
| chr11_85917986_85918561 | 0.16 |
Tbx4 |
T-box 4 |
21508 |
0.16 |
| chr4_13242090_13242241 | 0.16 |
Gm26250 |
predicted gene, 26250 |
24885 |
0.26 |
| chr1_38545635_38545840 | 0.15 |
Gm34727 |
predicted gene, 34727 |
58218 |
0.13 |
| chr7_136342313_136342682 | 0.15 |
C230079O03Rik |
RIKEN cDNA C230079O03 gene |
6918 |
0.21 |
| chr18_81091160_81091406 | 0.15 |
Gm50423 |
predicted gene, 50423 |
8219 |
0.2 |
| chr3_37906898_37907049 | 0.15 |
Gm20755 |
predicted gene, 20755 |
8160 |
0.18 |
| chr4_106977384_106977540 | 0.15 |
Ssbp3 |
single-stranded DNA binding protein 3 |
11036 |
0.19 |
| chr7_27375253_27375939 | 0.14 |
Gm20479 |
predicted gene 20479 |
18177 |
0.1 |
| chr2_76029030_76030067 | 0.13 |
Pde11a |
phosphodiesterase 11A |
23505 |
0.18 |
| chr11_57616045_57616196 | 0.13 |
Galnt10 |
polypeptide N-acetylgalactosaminyltransferase 10 |
29322 |
0.13 |
| chr13_23571443_23572971 | 0.13 |
H2bc8 |
H2B clustered histone 8 |
811 |
0.19 |
| chr11_49246951_49247357 | 0.13 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
323 |
0.83 |
| chr11_83877738_83877889 | 0.13 |
Hnf1b |
HNF1 homeobox B |
24853 |
0.13 |
| chr17_24959159_24959310 | 0.12 |
Jpt2 |
Jupiter microtubule associated homolog 2 |
1402 |
0.28 |
| chr10_60933986_60934521 | 0.12 |
Gm26947 |
predicted gene, 26947 |
2262 |
0.22 |
| chr1_172218328_172218648 | 0.12 |
Casq1 |
calsequestrin 1 |
1227 |
0.28 |
| chr10_77164464_77164999 | 0.12 |
Col18a1 |
collagen, type XVIII, alpha 1 |
1817 |
0.35 |
| chr5_111893894_111894510 | 0.12 |
Gm42488 |
predicted gene 42488 |
50033 |
0.15 |
| chr2_58000565_58000896 | 0.12 |
Galnt5 |
polypeptide N-acetylgalactosaminyltransferase 5 |
2846 |
0.28 |
| chr3_109574671_109575218 | 0.12 |
Vav3 |
vav 3 oncogene |
1037 |
0.69 |
| chr17_47143536_47144571 | 0.11 |
Trerf1 |
transcriptional regulating factor 1 |
2837 |
0.31 |
| chr2_72146177_72146373 | 0.11 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
2521 |
0.29 |
| chr16_21392882_21393033 | 0.11 |
Vps8 |
VPS8 CORVET complex subunit |
30161 |
0.18 |
| chr16_94714721_94714872 | 0.11 |
Gm41504 |
predicted gene, 41504 |
893 |
0.59 |
| chr9_60518348_60518989 | 0.11 |
Thsd4 |
thrombospondin, type I, domain containing 4 |
3378 |
0.18 |
| chr5_45534202_45534749 | 0.11 |
Fam184b |
family with sequence similarity 184, member B |
2067 |
0.2 |
| chr16_72665443_72666078 | 0.11 |
Robo1 |
roundabout guidance receptor 1 |
2556 |
0.45 |
| chr2_132562790_132562941 | 0.11 |
Gpcpd1 |
glycerophosphocholine phosphodiesterase 1 |
15268 |
0.16 |
| chr17_32318505_32319028 | 0.11 |
Akap8 |
A kinase (PRKA) anchor protein 8 |
38 |
0.96 |
| chr2_173789994_173790513 | 0.11 |
Vapb |
vesicle-associated membrane protein, associated protein B and C |
22053 |
0.16 |
| chr7_98721638_98721789 | 0.10 |
Gm19656 |
predicted gene, 19656 |
7047 |
0.13 |
| chr2_9906124_9906915 | 0.10 |
Gm13262 |
predicted gene 13262 |
341 |
0.81 |
| chr4_9831058_9831848 | 0.10 |
Gdf6 |
growth differentiation factor 6 |
12919 |
0.23 |
| chr10_122887302_122887925 | 0.10 |
Ppm1h |
protein phosphatase 1H (PP2C domain containing) |
7755 |
0.23 |
| chr2_160537495_160537646 | 0.10 |
Gm14222 |
predicted gene 14222 |
45787 |
0.15 |
| chr12_36826817_36826968 | 0.10 |
Mir5099 |
microRNA 5099 |
10687 |
0.24 |
| chr2_106822583_106822852 | 0.10 |
Gm22813 |
predicted gene, 22813 |
13381 |
0.23 |
| chr7_30908216_30909253 | 0.10 |
Mag |
myelin-associated glycoprotein |
5574 |
0.07 |
| chr9_60562470_60563218 | 0.10 |
Gm17853 |
predicted gene, 17853 |
14649 |
0.13 |
| chr11_108336593_108336967 | 0.10 |
Apoh |
apolipoprotein H |
6574 |
0.19 |
| chr5_149353036_149353786 | 0.10 |
BC028471 |
cDNA sequence BC028471 |
14225 |
0.11 |
| chr2_49272499_49272844 | 0.10 |
Mbd5 |
methyl-CpG binding domain protein 5 |
487 |
0.83 |
| chr5_93097700_93097987 | 0.10 |
Septin11 |
septin 11 |
4272 |
0.14 |
| chr6_107013042_107013673 | 0.10 |
Gm22418 |
predicted gene, 22418 |
11905 |
0.28 |
| chr9_41272710_41272970 | 0.09 |
Gm48710 |
predicted gene, 48710 |
43958 |
0.11 |
| chr7_89423254_89423405 | 0.09 |
AI314278 |
expressed sequence AI314278 |
2977 |
0.2 |
| chr19_17489963_17490448 | 0.09 |
Rfk |
riboflavin kinase |
92768 |
0.08 |
| chr18_54983558_54984280 | 0.09 |
Zfp608 |
zinc finger protein 608 |
6247 |
0.21 |
| chr2_90892674_90892825 | 0.09 |
Gm32514 |
predicted gene, 32514 |
449 |
0.68 |
| chr11_88313718_88313869 | 0.09 |
Ccdc182 |
coiled-coil domain containing 182 |
19735 |
0.14 |
| chr1_12695048_12695280 | 0.09 |
Sulf1 |
sulfatase 1 |
2632 |
0.26 |
| chr9_15308839_15309818 | 0.09 |
Taf1d |
TATA-box binding protein associated factor, RNA polymerase I, D |
572 |
0.37 |
| chr14_79530531_79530770 | 0.09 |
Elf1 |
E74-like factor 1 |
14952 |
0.15 |
| chr5_97935061_97936082 | 0.09 |
Antxr2 |
anthrax toxin receptor 2 |
60524 |
0.11 |
| chr14_27044544_27045365 | 0.09 |
Il17rd |
interleukin 17 receptor D |
5698 |
0.22 |
| chr10_8213906_8214393 | 0.09 |
Gm30906 |
predicted gene, 30906 |
38018 |
0.2 |
| chr9_110676440_110676899 | 0.09 |
Gm35715 |
predicted gene, 35715 |
1715 |
0.22 |
| chr2_157974620_157975884 | 0.09 |
Tti1 |
TELO2 interacting protein 1 |
31960 |
0.14 |
| chr1_88668363_88668968 | 0.09 |
Gm29336 |
predicted gene 29336 |
12357 |
0.16 |
| chr6_37645617_37646179 | 0.09 |
Ybx1-ps2 |
Y box protein 1, pseudogene 2 |
41297 |
0.18 |
| chr5_128684296_128685182 | 0.09 |
Piwil1 |
piwi-like RNA-mediated gene silencing 1 |
17785 |
0.17 |
| chr4_14103504_14103655 | 0.08 |
Gm24068 |
predicted gene, 24068 |
23639 |
0.22 |
| chr2_74696421_74697223 | 0.08 |
Gm28793 |
predicted gene 28793 |
764 |
0.23 |
| chr11_120710545_120710942 | 0.08 |
Cenpx |
centromere protein X |
1240 |
0.16 |
| chr5_113279864_113280669 | 0.08 |
Sgsm1 |
small G protein signaling modulator 1 |
321 |
0.85 |
| chr5_146282063_146282214 | 0.08 |
Cdk8 |
cyclin-dependent kinase 8 |
4038 |
0.19 |
| chr1_118776754_118776905 | 0.08 |
Gm28467 |
predicted gene 28467 |
33307 |
0.18 |
| chr2_139608706_139609326 | 0.08 |
Gm27385 |
predicted gene, 27385 |
20789 |
0.22 |
| chr11_120228562_120229205 | 0.08 |
2900052L18Rik |
RIKEN cDNA 2900052L18 gene |
2702 |
0.14 |
| chr4_32952734_32952939 | 0.08 |
Ankrd6 |
ankyrin repeat domain 6 |
1995 |
0.26 |
| chr1_51697218_51698004 | 0.08 |
Gm28055 |
predicted gene 28055 |
289 |
0.93 |
| chr5_101323183_101323902 | 0.08 |
Cycs-ps2 |
cytochrome c, pseudogene 2 |
17964 |
0.27 |
| chr7_135526475_135527168 | 0.08 |
Clrn3 |
clarin 3 |
1833 |
0.31 |
| chr19_16168628_16168779 | 0.08 |
E030024N20Rik |
RIKEN cDNA E030024N20 gene |
3898 |
0.26 |
| chr14_22168600_22169219 | 0.08 |
Gm7480 |
predicted gene 7480 |
132806 |
0.04 |
| chr2_38914989_38915140 | 0.08 |
Nr6a1 |
nuclear receptor subfamily 6, group A, member 1 |
11153 |
0.11 |
| chr3_18241670_18242257 | 0.08 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
1375 |
0.5 |
| chr5_144098470_144098725 | 0.08 |
Lmtk2 |
lemur tyrosine kinase 2 |
1839 |
0.26 |
| chr10_22813360_22814229 | 0.08 |
Gm10824 |
predicted gene 10824 |
1955 |
0.28 |
| chr15_27998794_27999022 | 0.07 |
Trio |
triple functional domain (PTPRF interacting) |
3545 |
0.28 |
| chr3_30514092_30514862 | 0.07 |
Mecom |
MDS1 and EVI1 complex locus |
4655 |
0.16 |
| chr13_78194785_78195954 | 0.07 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
1004 |
0.42 |
| chr5_148820568_148820913 | 0.07 |
Gm15407 |
predicted gene 15407 |
27 |
0.97 |
| chr18_15060481_15060964 | 0.07 |
Kctd1 |
potassium channel tetramerisation domain containing 1 |
1167 |
0.55 |
| chr5_66503946_66504664 | 0.07 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
10525 |
0.15 |
| chr18_60646141_60646566 | 0.07 |
Synpo |
synaptopodin |
1919 |
0.32 |
| chr19_14777221_14777986 | 0.07 |
Gm26026 |
predicted gene, 26026 |
59123 |
0.15 |
| chr16_38458401_38458552 | 0.07 |
Cd80 |
CD80 antigen |
457 |
0.72 |
| chr1_79823877_79824635 | 0.07 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
2934 |
0.26 |
| chr11_96863722_96864075 | 0.07 |
Gm11524 |
predicted gene 11524 |
6297 |
0.09 |
| chr2_152753689_152753840 | 0.07 |
Cox4i2 |
cytochrome c oxidase subunit 4I2 |
409 |
0.74 |
| chr10_45470864_45471317 | 0.07 |
Lin28b |
lin-28 homolog B (C. elegans) |
889 |
0.65 |
| chr10_78752230_78753214 | 0.07 |
Ccdc105 |
coiled-coil domain containing 105 |
343 |
0.81 |
| chr11_63917212_63917744 | 0.07 |
Hs3st3b1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
4812 |
0.25 |
| chr7_66449827_66450975 | 0.07 |
Gm44753 |
predicted gene 44753 |
8515 |
0.17 |
| chr13_63276050_63276513 | 0.07 |
Aopep |
aminopeptidase O |
2389 |
0.12 |
| chr3_107943213_107944057 | 0.07 |
Gstm6 |
glutathione S-transferase, mu 6 |
80 |
0.92 |
| chr8_11311242_11311393 | 0.07 |
Col4a1 |
collagen, type IV, alpha 1 |
1372 |
0.31 |
| chr11_5972524_5972675 | 0.07 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
11539 |
0.13 |
| chr2_170070487_170071222 | 0.06 |
Zfp217 |
zinc finger protein 217 |
60366 |
0.14 |
| chr10_41292796_41293533 | 0.06 |
Fig4 |
FIG4 phosphoinositide 5-phosphatase |
10096 |
0.16 |
| chr1_167374494_167374876 | 0.06 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
10519 |
0.13 |
| chr12_95547341_95547492 | 0.06 |
Gm22246 |
predicted gene, 22246 |
72402 |
0.11 |
| chr2_91529031_91529182 | 0.06 |
Ckap5 |
cytoskeleton associated protein 5 |
2128 |
0.25 |
| chr10_51554019_51554385 | 0.06 |
Gm48787 |
predicted gene, 48787 |
2755 |
0.15 |
| chr8_23413505_23413993 | 0.06 |
Sfrp1 |
secreted frizzled-related protein 1 |
2247 |
0.39 |
| chr4_149605840_149605991 | 0.06 |
Clstn1 |
calsyntenin 1 |
18667 |
0.12 |
| chr16_94180124_94180691 | 0.06 |
Hlcs |
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
5407 |
0.13 |
| chr2_174283563_174287177 | 0.06 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
12 |
0.53 |
| chr11_76672356_76673645 | 0.06 |
Bhlha9 |
basic helix-loop-helix family, member a9 |
530 |
0.76 |
| chr11_96876203_96877056 | 0.06 |
Gm11523 |
predicted gene 11523 |
2675 |
0.13 |
| chrX_141726225_141726376 | 0.06 |
Irs4 |
insulin receptor substrate 4 |
1037 |
0.41 |
| chr8_27162795_27163172 | 0.06 |
Gm45267 |
predicted gene 45267 |
3155 |
0.13 |
| chr5_136051364_136052209 | 0.06 |
Upk3bl |
uroplakin 3B-like |
2706 |
0.15 |
| chr11_70404123_70404274 | 0.06 |
Pelp1 |
proline, glutamic acid and leucine rich protein 1 |
5830 |
0.09 |
| chr14_52306804_52306955 | 0.06 |
Mettl3 |
methyltransferase like 3 |
1751 |
0.18 |
| chr8_125361611_125361762 | 0.06 |
Sipa1l2 |
signal-induced proliferation-associated 1 like 2 |
131024 |
0.05 |
| chr17_48039617_48039969 | 0.06 |
1700122O11Rik |
RIKEN cDNA 1700122O11 gene |
1500 |
0.3 |
| chr15_50883181_50884101 | 0.06 |
Trps1 |
transcriptional repressor GATA binding 1 |
835 |
0.64 |
| chrX_133689241_133690268 | 0.06 |
Pcdh19 |
protocadherin 19 |
767 |
0.77 |
| chr3_95517184_95517335 | 0.06 |
Ctss |
cathepsin S |
9527 |
0.1 |
| chr17_47989917_47990574 | 0.06 |
Gm14871 |
predicted gene 14871 |
13327 |
0.14 |
| chr19_28580015_28580320 | 0.06 |
4933413C19Rik |
RIKEN cDNA 4933413C19 gene |
3127 |
0.3 |
| chr18_3666230_3667225 | 0.06 |
Gm50091 |
predicted gene, 50091 |
50446 |
0.14 |
| chr17_43758947_43759124 | 0.06 |
Cyp39a1 |
cytochrome P450, family 39, subfamily a, polypeptide 1 |
27092 |
0.17 |
| chr1_91762985_91763136 | 0.06 |
Twist2 |
twist basic helix-loop-helix transcription factor 2 |
38401 |
0.14 |
| chr8_124990881_124991137 | 0.06 |
Tsnax |
translin-associated factor X |
21988 |
0.13 |
| chr12_59039859_59040384 | 0.06 |
Gm22973 |
predicted gene, 22973 |
214 |
0.89 |
| chr18_55258362_55259199 | 0.06 |
Gm22597 |
predicted gene, 22597 |
78940 |
0.09 |
| chr6_91712146_91712297 | 0.06 |
Gm44981 |
predicted gene 44981 |
1057 |
0.39 |
| chr14_21988628_21988808 | 0.06 |
Zfp503 |
zinc finger protein 503 |
883 |
0.5 |
| chr18_74966819_74967388 | 0.06 |
Lipg |
lipase, endothelial |
5840 |
0.1 |
| chr2_162967542_162967693 | 0.06 |
Gm11453 |
predicted gene 11453 |
15644 |
0.11 |
| chr18_50046880_50047192 | 0.06 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
296 |
0.91 |
| chr9_56842487_56842638 | 0.06 |
Odf3l1 |
outer dense fiber of sperm tails 3-like 1 |
9401 |
0.14 |
| chr12_107960709_107960860 | 0.06 |
Bcl11b |
B cell leukemia/lymphoma 11B |
42630 |
0.19 |
| chr9_98538219_98538951 | 0.06 |
4930579K19Rik |
RIKEN cDNA 4930579K19 gene |
25007 |
0.11 |
| chr5_75975002_75975424 | 0.06 |
Kdr |
kinase insert domain protein receptor |
3245 |
0.23 |
| chr8_26384770_26384921 | 0.06 |
Gm31898 |
predicted gene, 31898 |
50868 |
0.1 |
| chr11_18872620_18872876 | 0.06 |
8430419K02Rik |
RIKEN cDNA 8430419K02 gene |
593 |
0.71 |
| chr3_51357869_51359319 | 0.06 |
Gm37203 |
predicted gene, 37203 |
11628 |
0.1 |
| chr14_118322252_118323294 | 0.06 |
Gm32093 |
predicted gene, 32093 |
204 |
0.92 |
| chr10_125839451_125839817 | 0.06 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
126534 |
0.06 |
| chr18_55106919_55107352 | 0.06 |
AC163347.1 |
novel transcript |
10775 |
0.17 |
| chr4_116688529_116688680 | 0.06 |
Prdx1 |
peroxiredoxin 1 |
1292 |
0.29 |
| chr5_33534271_33535601 | 0.06 |
Fam53a |
family with sequence similarity 53, member A |
93976 |
0.05 |
| chr15_36779468_36779639 | 0.06 |
Ywhaz |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
12861 |
0.15 |
| chr4_66414553_66415137 | 0.06 |
Astn2 |
astrotactin 2 |
10234 |
0.28 |
| chr14_63255547_63256547 | 0.06 |
Gata4 |
GATA binding protein 4 |
10776 |
0.17 |
| chr17_29080873_29081286 | 0.06 |
Trp53cor1 |
tumor protein p53 pathway corepressor 1 |
1900 |
0.17 |
| chr17_29397536_29397687 | 0.06 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
21110 |
0.11 |
| chr5_92811720_92812319 | 0.06 |
Shroom3 |
shroom family member 3 |
2549 |
0.29 |
| chr1_183144103_183144344 | 0.06 |
Disp1 |
dispatched RND transporter family member 1 |
3238 |
0.28 |
| chr3_118548450_118548601 | 0.06 |
Gm32444 |
predicted gene, 32444 |
11504 |
0.15 |
| chr1_41131507_41131710 | 0.05 |
4930448I06Rik |
RIKEN cDNA 4930448I06 gene |
49644 |
0.19 |
| chr11_35972396_35972620 | 0.05 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
8019 |
0.23 |
| chr1_87175864_87176092 | 0.05 |
Prss56 |
protease, serine 56 |
7335 |
0.1 |
| chr4_148890637_148891853 | 0.05 |
Casz1 |
castor zinc finger 1 |
1864 |
0.33 |
| chr3_41522196_41522994 | 0.05 |
2400006E01Rik |
RIKEN cDNA 2400006E01 gene |
622 |
0.65 |
| chr6_117886217_117886368 | 0.05 |
Gm29509 |
predicted gene 29509 |
12909 |
0.11 |
| chr10_116276295_116276851 | 0.05 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
1023 |
0.54 |
| chr2_165494605_165494756 | 0.05 |
Trp53rka |
transformation related protein 53 regulating kinase A |
1356 |
0.36 |
| chr8_26308366_26308822 | 0.05 |
Gm31784 |
predicted gene, 31784 |
3740 |
0.18 |
| chr9_54769552_54770224 | 0.05 |
Crabp1 |
cellular retinoic acid binding protein I |
5140 |
0.18 |
| chr3_41284805_41285284 | 0.05 |
Gm16508 |
predicted gene 16508 |
5546 |
0.25 |
| chr1_106602971_106603597 | 0.05 |
Gm37053 |
predicted gene, 37053 |
3586 |
0.27 |
| chr6_51002449_51002688 | 0.05 |
Gm44402 |
predicted gene, 44402 |
35300 |
0.18 |
| chr11_69549158_69549411 | 0.05 |
Dnah2 |
dynein, axonemal, heavy chain 2 |
174 |
0.88 |
| chr5_104109155_104109917 | 0.05 |
Gm26703 |
predicted gene, 26703 |
245 |
0.88 |
| chr9_60279898_60280278 | 0.05 |
2010001M07Rik |
RIKEN cDNA 2010001M07 gene |
54054 |
0.14 |
| chr10_100050584_100050771 | 0.05 |
Kitl |
kit ligand |
34761 |
0.14 |
| chr17_91954712_91954863 | 0.05 |
Gm41654 |
predicted gene, 41654 |
82648 |
0.09 |
| chr10_45808506_45808657 | 0.05 |
D030045P18Rik |
RIKEN cDNA D030045P18 gene |
877 |
0.63 |
| chr9_106397846_106397997 | 0.05 |
Dusp7 |
dual specificity phosphatase 7 |
23302 |
0.09 |
| chr11_97047163_97047404 | 0.05 |
Osbpl7 |
oxysterol binding protein-like 7 |
3345 |
0.09 |
| chrX_7422152_7422543 | 0.05 |
2010204K13Rik |
RIKEN cDNA 2010204K13 gene |
641 |
0.67 |
| chr19_56631461_56631906 | 0.05 |
Gm32441 |
predicted gene, 32441 |
32928 |
0.14 |
| chr2_95232237_95232956 | 0.05 |
Gm13794 |
predicted gene 13794 |
161646 |
0.04 |
| chr8_26808819_26808970 | 0.05 |
2310008N11Rik |
RIKEN cDNA 2310008N11 gene |
15529 |
0.17 |
| chr12_27566196_27567004 | 0.05 |
Gm4166 |
predicted gene 4166 |
31601 |
0.22 |
| chr1_89848531_89848837 | 0.05 |
Gm37521 |
predicted gene, 37521 |
23203 |
0.19 |
| chr4_42183593_42183996 | 0.05 |
1700045I11Rik |
RIKEN cDNA 1700045I11 gene |
12949 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0048369 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |