Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
Results for Hoxc10
Z-value: 0.43
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSMUSG00000022484.7 | homeobox C10 |
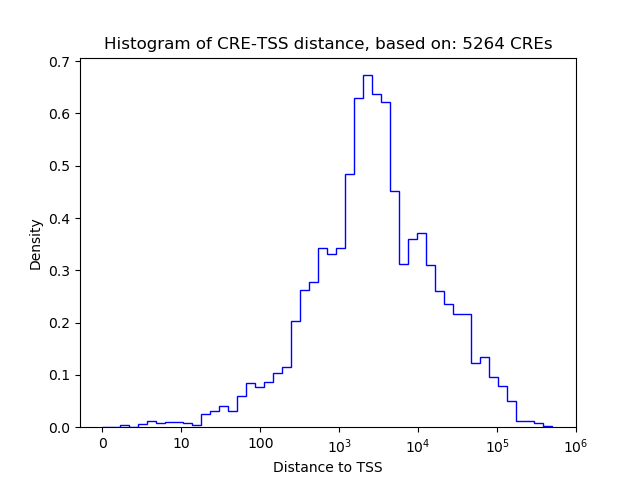
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_102964234_102964441 | Hoxc10 | 2459 | 0.118754 | -0.19 | 1.5e-01 | Click! |
| chr15_102961468_102962108 | Hoxc10 | 5008 | 0.085565 | -0.18 | 1.7e-01 | Click! |
| chr15_102964487_102964867 | Hoxc10 | 2119 | 0.132689 | -0.17 | 1.8e-01 | Click! |
| chr15_102962399_102963013 | Hoxc10 | 4090 | 0.091179 | -0.15 | 2.7e-01 | Click! |
| chr15_102965066_102967702 | Hoxc10 | 412 | 0.596726 | -0.14 | 2.7e-01 | Click! |
Activity of the Hoxc10 motif across conditions
Conditions sorted by the z-value of the Hoxc10 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
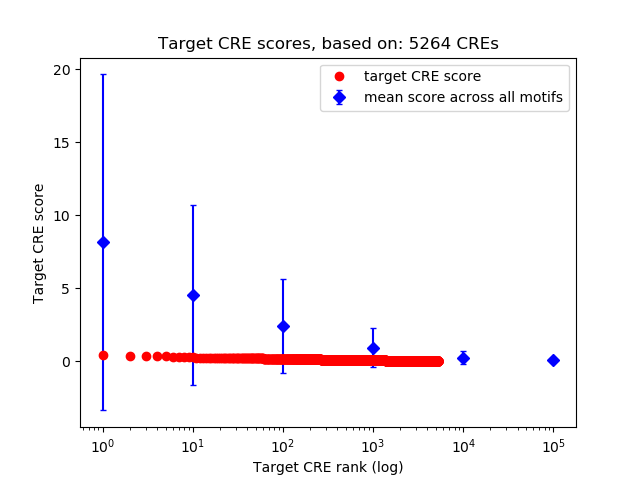
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_41702194_41702736 | 0.40 |
Kel |
Kell blood group |
1874 |
0.23 |
| chr9_91363064_91363717 | 0.38 |
Zic1 |
zinc finger protein of the cerebellum 1 |
336 |
0.72 |
| chr8_12399326_12400483 | 0.37 |
Gm25239 |
predicted gene, 25239 |
3501 |
0.16 |
| chr5_116589538_116590511 | 0.36 |
Srrm4 |
serine/arginine repetitive matrix 4 |
1793 |
0.34 |
| chr2_71536976_71537311 | 0.36 |
Dlx1as |
distal-less homeobox 1, antisense |
667 |
0.61 |
| chr12_52699339_52699808 | 0.29 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
190 |
0.95 |
| chr12_118850924_118851621 | 0.29 |
Sp8 |
trans-acting transcription factor 8 |
3686 |
0.27 |
| chr10_92160735_92161461 | 0.28 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
1663 |
0.4 |
| chr5_98180058_98181966 | 0.27 |
Prdm8 |
PR domain containing 8 |
34 |
0.97 |
| chr3_96248416_96249149 | 0.27 |
H3c14 |
H3 clustered histone 14 |
2097 |
0.07 |
| chr13_83727321_83728283 | 0.26 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
304 |
0.83 |
| chr6_83315880_83316808 | 0.26 |
Gm43890 |
predicted gene, 43890 |
599 |
0.53 |
| chr11_109614897_109615494 | 0.25 |
Gm11685 |
predicted gene 11685 |
1125 |
0.43 |
| chr9_91350199_91351559 | 0.25 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
74 |
0.95 |
| chr13_78178635_78180138 | 0.25 |
Gm38604 |
predicted gene, 38604 |
3773 |
0.16 |
| chr1_131135705_131136011 | 0.25 |
Dyrk3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
2387 |
0.21 |
| chr2_38342005_38342312 | 0.24 |
Lhx2 |
LIM homeobox protein 2 |
1066 |
0.44 |
| chr2_170192748_170194111 | 0.24 |
Zfp217 |
zinc finger protein 217 |
45326 |
0.17 |
| chr4_74300833_74301004 | 0.24 |
Kdm4c |
lysine (K)-specific demethylase 4C |
38108 |
0.16 |
| chr8_80496570_80496939 | 0.24 |
Gypa |
glycophorin A |
2973 |
0.3 |
| chr4_96346915_96347066 | 0.23 |
Cyp2j11 |
cytochrome P450, family 2, subfamily j, polypeptide 11 |
1672 |
0.4 |
| chr5_8063599_8063897 | 0.23 |
Sri |
sorcin |
2482 |
0.22 |
| chr6_39418262_39418473 | 0.23 |
Mkrn1 |
makorin, ring finger protein, 1 |
1390 |
0.27 |
| chr14_12189998_12191323 | 0.23 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
717 |
0.71 |
| chr3_88206822_88208169 | 0.23 |
Gm3764 |
predicted gene 3764 |
183 |
0.86 |
| chr2_22625054_22625976 | 0.22 |
Gad2 |
glutamic acid decarboxylase 2 |
2211 |
0.22 |
| chr9_91360032_91360505 | 0.22 |
Zic4 |
zinc finger protein of the cerebellum 4 |
2145 |
0.17 |
| chr16_35810345_35810496 | 0.22 |
Gm26838 |
predicted gene, 26838 |
1474 |
0.31 |
| chr3_17790150_17790808 | 0.22 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
522 |
0.77 |
| chr14_55061871_55064122 | 0.22 |
Gm20687 |
predicted gene 20687 |
7503 |
0.08 |
| chr3_17793389_17793789 | 0.22 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
331 |
0.86 |
| chr1_26388093_26388357 | 0.22 |
Gm19123 |
predicted gene, 19123 |
78011 |
0.12 |
| chr16_18429039_18430122 | 0.22 |
Txnrd2 |
thioredoxin reductase 2 |
655 |
0.54 |
| chr3_141989393_141989691 | 0.22 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
7278 |
0.31 |
| chr10_21695137_21695288 | 0.22 |
Gm5420 |
predicted gene 5420 |
4367 |
0.25 |
| chr17_66869630_66870110 | 0.21 |
Gm49940 |
predicted gene, 49940 |
7630 |
0.18 |
| chr13_99493273_99493585 | 0.21 |
5330431K02Rik |
RIKEN cDNA 5330431K02 gene |
11010 |
0.16 |
| chr2_31638722_31641540 | 0.21 |
Prdm12 |
PR domain containing 12 |
94 |
0.84 |
| chr1_57083098_57083538 | 0.21 |
9130024F11Rik |
RIKEN cDNA 9130024F11 gene |
42925 |
0.14 |
| chr1_40771505_40772300 | 0.21 |
Gm37915 |
predicted gene, 37915 |
517 |
0.75 |
| chr8_41052368_41053980 | 0.21 |
Gm16193 |
predicted gene 16193 |
64 |
0.96 |
| chr17_46485045_46485196 | 0.21 |
Ttbk1 |
tau tubulin kinase 1 |
825 |
0.43 |
| chr11_20330074_20330777 | 0.21 |
Slc1a4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
2259 |
0.28 |
| chr1_25228097_25229399 | 0.21 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
78 |
0.96 |
| chr3_104632268_104633005 | 0.21 |
Gm43582 |
predicted gene 43582 |
2021 |
0.18 |
| chr6_34869198_34869349 | 0.21 |
Tmem140 |
transmembrane protein 140 |
2221 |
0.2 |
| chrX_166346283_166346827 | 0.21 |
Gpm6b |
glycoprotein m6b |
1713 |
0.43 |
| chr4_23636118_23636607 | 0.21 |
Gm25978 |
predicted gene, 25978 |
9617 |
0.24 |
| chr3_121460231_121460981 | 0.20 |
Slc44a3 |
solute carrier family 44, member 3 |
305 |
0.86 |
| chr18_34519817_34520312 | 0.20 |
n-R5s24 |
nuclear encoded rRNA 5S 24 |
10327 |
0.14 |
| chr4_46453910_46454934 | 0.20 |
Anp32b |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
3520 |
0.17 |
| chr4_48050018_48050169 | 0.20 |
Nr4a3 |
nuclear receptor subfamily 4, group A, member 3 |
979 |
0.62 |
| chr13_83749435_83750073 | 0.20 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
10891 |
0.12 |
| chr1_95665138_95666758 | 0.20 |
St8sia4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
1587 |
0.41 |
| chr18_70573145_70573296 | 0.20 |
Mbd2 |
methyl-CpG binding domain protein 2 |
4886 |
0.22 |
| chr18_25751839_25752425 | 0.19 |
Celf4 |
CUGBP, Elav-like family member 4 |
560 |
0.81 |
| chr7_70347472_70349327 | 0.19 |
Gm44948 |
predicted gene 44948 |
703 |
0.54 |
| chr1_133755015_133755406 | 0.19 |
Atp2b4 |
ATPase, Ca++ transporting, plasma membrane 4 |
1463 |
0.34 |
| chr4_125727107_125727874 | 0.19 |
2610028E06Rik |
RIKEN cDNA 2610028E06 gene |
171492 |
0.03 |
| chr4_110285468_110287125 | 0.19 |
Elavl4 |
ELAV like RNA binding protein 4 |
320 |
0.94 |
| chr9_19302176_19302327 | 0.19 |
Olfr844 |
olfactory receptor 844 |
14096 |
0.13 |
| chr13_23451039_23451496 | 0.19 |
Gm46404 |
predicted gene, 46404 |
5332 |
0.09 |
| chr16_35809219_35809489 | 0.19 |
Gm26838 |
predicted gene, 26838 |
408 |
0.78 |
| chr1_58960528_58960716 | 0.19 |
Trak2 |
trafficking protein, kinesin binding 2 |
12807 |
0.14 |
| chr1_42688699_42689514 | 0.19 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
3987 |
0.16 |
| chr8_84768788_84768967 | 0.19 |
Nfix |
nuclear factor I/X |
4519 |
0.13 |
| chr10_14703599_14704234 | 0.19 |
Vta1 |
vesicle (multivesicular body) trafficking 1 |
1543 |
0.26 |
| chr9_48338929_48340200 | 0.19 |
Nxpe2 |
neurexophilin and PC-esterase domain family, member 2 |
1270 |
0.48 |
| chr1_162866141_162866550 | 0.18 |
Fmo1 |
flavin containing monooxygenase 1 |
265 |
0.91 |
| chr8_47346057_47346419 | 0.18 |
Stox2 |
storkhead box 2 |
6110 |
0.26 |
| chr16_81285172_81285323 | 0.18 |
Gm49555 |
predicted gene, 49555 |
4915 |
0.27 |
| chr2_105678552_105679922 | 0.18 |
Pax6 |
paired box 6 |
630 |
0.68 |
| chr10_103026340_103027824 | 0.18 |
Alx1 |
ALX homeobox 1 |
1545 |
0.39 |
| chr8_123858032_123858183 | 0.18 |
Ccsap |
centriole, cilia and spindle associated protein |
1423 |
0.23 |
| chr1_177448882_177449429 | 0.18 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
3334 |
0.19 |
| chr14_69278760_69279066 | 0.18 |
Gm20236 |
predicted gene, 20236 |
3227 |
0.12 |
| chrX_110811626_110812334 | 0.18 |
Gm44593 |
predicted gene 44593 |
344 |
0.89 |
| chr8_12400578_12402091 | 0.18 |
Gm25239 |
predicted gene, 25239 |
4931 |
0.15 |
| chr10_67002257_67005140 | 0.18 |
Gm31763 |
predicted gene, 31763 |
1322 |
0.45 |
| chr10_94020545_94020760 | 0.18 |
Vezt |
vezatin, adherens junctions transmembrane protein |
71 |
0.96 |
| chr2_28620746_28622145 | 0.18 |
Gfi1b |
growth factor independent 1B |
500 |
0.68 |
| chr19_31028064_31028215 | 0.18 |
Cstf2t |
cleavage stimulation factor, 3' pre-RNA subunit 2, tau |
54698 |
0.16 |
| chr14_122480308_122481080 | 0.18 |
Zic2 |
zinc finger protein of the cerebellum 2 |
2594 |
0.16 |
| chr6_113890285_113891668 | 0.18 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
394 |
0.85 |
| chr7_121070927_121071078 | 0.18 |
Igsf6 |
immunoglobulin superfamily, member 6 |
3570 |
0.11 |
| chr4_100855622_100855877 | 0.18 |
Cachd1 |
cache domain containing 1 |
79074 |
0.1 |
| chr11_32290084_32290341 | 0.17 |
Hbq1b |
hemoglobin, theta 1B |
3211 |
0.13 |
| chr9_77994297_77994513 | 0.17 |
Gm34829 |
predicted gene, 34829 |
1806 |
0.24 |
| chr14_32146045_32146369 | 0.17 |
Msmb |
beta-microseminoprotein |
1380 |
0.29 |
| chr16_77422348_77423278 | 0.17 |
9430053O09Rik |
RIKEN cDNA 9430053O09 gene |
993 |
0.4 |
| chr17_3482528_3483679 | 0.17 |
Tiam2 |
T cell lymphoma invasion and metastasis 2 |
387 |
0.86 |
| chr5_137024960_137025964 | 0.17 |
Vgf |
VGF nerve growth factor inducible |
930 |
0.37 |
| chr1_42701498_42701775 | 0.17 |
Pou3f3 |
POU domain, class 3, transcription factor 3 |
5868 |
0.14 |
| chr12_53061586_53062872 | 0.17 |
n-R5s58 |
nuclear encoded rRNA 5S 58 |
15403 |
0.29 |
| chr2_61806407_61806964 | 0.17 |
Tbr1 |
T-box brain gene 1 |
412 |
0.83 |
| chr13_9095816_9096692 | 0.17 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
2272 |
0.25 |
| chr9_91363965_91365514 | 0.17 |
Zic1 |
zinc finger protein of the cerebellum 1 |
1029 |
0.35 |
| chr3_129836696_129837785 | 0.17 |
Cfi |
complement component factor i |
503 |
0.7 |
| chr19_59347278_59347441 | 0.17 |
Pdzd8 |
PDZ domain containing 8 |
1579 |
0.32 |
| chr9_74865733_74868961 | 0.17 |
Onecut1 |
one cut domain, family member 1 |
863 |
0.54 |
| chr19_56543836_56544364 | 0.17 |
Dclre1a |
DNA cross-link repair 1A |
2555 |
0.26 |
| chr7_27735092_27735839 | 0.17 |
Zfp60 |
zinc finger protein 60 |
902 |
0.47 |
| chr3_34656299_34657721 | 0.17 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
974 |
0.4 |
| chr11_111996362_111996587 | 0.17 |
Gm11679 |
predicted gene 11679 |
47104 |
0.19 |
| chr4_53633406_53634189 | 0.17 |
Fsd1l |
fibronectin type III and SPRY domain containing 1-like |
2081 |
0.3 |
| chr16_43509882_43510133 | 0.17 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
301 |
0.93 |
| chr10_26230524_26230675 | 0.17 |
Samd3 |
sterile alpha motif domain containing 3 |
3 |
0.98 |
| chr9_66276261_66276412 | 0.17 |
Dapk2 |
death-associated protein kinase 2 |
7962 |
0.23 |
| chr11_96347886_96350398 | 0.17 |
Hoxb3os |
homeobox B3 and homeobox B2, opposite strand |
1219 |
0.21 |
| chr11_98708998_98709149 | 0.17 |
Med24 |
mediator complex subunit 24 |
1070 |
0.29 |
| chr2_48538709_48539576 | 0.16 |
Gm13481 |
predicted gene 13481 |
81897 |
0.1 |
| chr15_77953936_77954120 | 0.16 |
Foxred2 |
FAD-dependent oxidoreductase domain containing 2 |
2647 |
0.2 |
| chr1_42242281_42242432 | 0.16 |
Gm29664 |
predicted gene 29664 |
3661 |
0.22 |
| chr8_85378394_85378546 | 0.16 |
Mylk3 |
myosin light chain kinase 3 |
2508 |
0.21 |
| chr17_25090137_25090691 | 0.16 |
Ift140 |
intraflagellar transport 140 |
257 |
0.85 |
| chr1_142984549_142984700 | 0.16 |
Gm5835 |
predicted gene 5835 |
109992 |
0.07 |
| chr19_28677931_28680368 | 0.16 |
Glis3 |
GLIS family zinc finger 3 |
459 |
0.68 |
| chr10_57787071_57787893 | 0.16 |
Fabp7 |
fatty acid binding protein 7, brain |
2559 |
0.25 |
| chr14_19974553_19975713 | 0.16 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
1560 |
0.43 |
| chr1_165766563_165766714 | 0.16 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
2838 |
0.13 |
| chr18_31446492_31447667 | 0.16 |
Syt4 |
synaptotagmin IV |
327 |
0.87 |
| chr1_42262366_42263014 | 0.16 |
Gm28175 |
predicted gene 28175 |
833 |
0.63 |
| chr16_30266391_30266905 | 0.16 |
Cpn2 |
carboxypeptidase N, polypeptide 2 |
851 |
0.52 |
| chr6_6857856_6858925 | 0.16 |
Gm44094 |
predicted gene, 44094 |
2139 |
0.2 |
| chr9_83253415_83254213 | 0.16 |
Gm27216 |
predicted gene 27216 |
208 |
0.95 |
| chr2_70567574_70569064 | 0.16 |
Gad1 |
glutamate decarboxylase 1 |
1942 |
0.26 |
| chr14_34623313_34624132 | 0.16 |
Opn4 |
opsin 4 (melanopsin) |
23580 |
0.11 |
| chr3_34562856_34563429 | 0.16 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
2750 |
0.22 |
| chr2_23037510_23037739 | 0.16 |
Abi1 |
abl-interactor 1 |
2480 |
0.26 |
| chr5_88587396_88587564 | 0.16 |
Rufy3 |
RUN and FYVE domain containing 3 |
3686 |
0.2 |
| chr2_52557337_52558561 | 0.16 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
611 |
0.74 |
| chr1_42230426_42230769 | 0.16 |
Gm9915 |
predicted gene 9915 |
870 |
0.59 |
| chr7_53564328_53564479 | 0.16 |
Mir6238 |
microRNA 6238 |
327486 |
0.01 |
| chr9_52145956_52147029 | 0.16 |
Zc3h12c |
zinc finger CCCH type containing 12C |
21619 |
0.17 |
| chr14_115043258_115044172 | 0.16 |
Mir17 |
microRNA 17 |
44 |
0.47 |
| chr13_83715222_83716973 | 0.16 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
5284 |
0.15 |
| chr3_56179928_56180616 | 0.16 |
Nbea |
neurobeachin |
3429 |
0.25 |
| chr12_49394720_49395481 | 0.16 |
3110039M20Rik |
RIKEN cDNA 3110039M20 gene |
4441 |
0.15 |
| chr9_91366433_91367646 | 0.15 |
Zic1 |
zinc finger protein of the cerebellum 1 |
1229 |
0.26 |
| chr2_105671719_105672950 | 0.15 |
Pax6 |
paired box 6 |
474 |
0.75 |
| chr10_119878178_119878329 | 0.15 |
Grip1os1 |
glutamate receptor interacting protein 1, opposite strand 1 |
25863 |
0.19 |
| chr15_66809584_66809797 | 0.15 |
Sla |
src-like adaptor |
2903 |
0.26 |
| chr1_178299267_178300366 | 0.15 |
Gm37486 |
predicted gene, 37486 |
420 |
0.74 |
| chr12_49390931_49392462 | 0.15 |
3110039M20Rik |
RIKEN cDNA 3110039M20 gene |
1037 |
0.42 |
| chr12_56532060_56532690 | 0.15 |
Nkx2-1 |
NK2 homeobox 1 |
2731 |
0.16 |
| chr6_38121806_38121957 | 0.15 |
Atp6v0a4 |
ATPase, H+ transporting, lysosomal V0 subunit A4 |
2608 |
0.21 |
| chr13_83742301_83742867 | 0.15 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
3721 |
0.15 |
| chr4_46401482_46401633 | 0.15 |
Hemgn |
hemogen |
2679 |
0.19 |
| chr2_16360798_16360949 | 0.15 |
Plxdc2 |
plexin domain containing 2 |
3756 |
0.39 |
| chr13_97250234_97250508 | 0.15 |
Enc1 |
ectodermal-neural cortex 1 |
9266 |
0.17 |
| chr18_39904089_39904240 | 0.15 |
Gm41708 |
predicted gene, 41708 |
25210 |
0.24 |
| chr4_100616357_100616508 | 0.15 |
Gm12700 |
predicted gene 12700 |
35154 |
0.17 |
| chr7_79505833_79506958 | 0.15 |
Mir9-3 |
microRNA 9-3 |
1131 |
0.28 |
| chr2_86873075_86873226 | 0.15 |
Gm13723 |
predicted gene 13723 |
1652 |
0.22 |
| chr11_78074087_78074838 | 0.15 |
Mir451b |
microRNA 451b |
1221 |
0.18 |
| chr1_168425831_168426144 | 0.15 |
Pbx1 |
pre B cell leukemia homeobox 1 |
5517 |
0.27 |
| chr14_60636814_60637375 | 0.15 |
Spata13 |
spermatogenesis associated 13 |
2339 |
0.35 |
| chr14_29807795_29808268 | 0.15 |
Gm35281 |
predicted gene, 35281 |
19266 |
0.19 |
| chr13_83717521_83718816 | 0.15 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
3213 |
0.17 |
| chr8_80495549_80496014 | 0.15 |
Gypa |
glycophorin A |
2000 |
0.38 |
| chr6_17750183_17750783 | 0.15 |
St7 |
suppression of tumorigenicity 7 |
1091 |
0.38 |
| chr9_14369210_14369409 | 0.15 |
Endod1 |
endonuclease domain containing 1 |
11674 |
0.12 |
| chr16_18430494_18430682 | 0.15 |
Txnrd2 |
thioredoxin reductase 2 |
1663 |
0.22 |
| chrX_166172119_166172805 | 0.15 |
Gemin8 |
gem nuclear organelle associated protein 8 |
1967 |
0.37 |
| chr9_113970028_113970753 | 0.15 |
Gm47950 |
predicted gene, 47950 |
608 |
0.61 |
| chr18_43686487_43688415 | 0.15 |
Jakmip2 |
janus kinase and microtubule interacting protein 2 |
174 |
0.96 |
| chr9_75232729_75232880 | 0.14 |
Myo5c |
myosin VC |
728 |
0.65 |
| chr6_15186533_15187348 | 0.14 |
Foxp2 |
forkhead box P2 |
1377 |
0.61 |
| chr10_42581935_42584872 | 0.14 |
Nr2e1 |
nuclear receptor subfamily 2, group E, member 1 |
229 |
0.69 |
| chr8_120507179_120507330 | 0.14 |
Gm26971 |
predicted gene, 26971 |
13361 |
0.12 |
| chr17_80801064_80802812 | 0.14 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
73453 |
0.09 |
| chr4_23601397_23602347 | 0.14 |
Gm25978 |
predicted gene, 25978 |
24873 |
0.22 |
| chr6_57822563_57823744 | 0.14 |
Vopp1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
1487 |
0.3 |
| chr2_138118544_138118695 | 0.14 |
Btbd3 |
BTB (POZ) domain containing 3 |
137946 |
0.05 |
| chr16_91397742_91397893 | 0.14 |
Gm21970 |
predicted gene 21970 |
6096 |
0.12 |
| chr8_41054160_41054452 | 0.14 |
Mtus1 |
mitochondrial tumor suppressor 1 |
477 |
0.69 |
| chr4_147814549_147814848 | 0.14 |
Gm20707 |
predicted gene 20707 |
4858 |
0.11 |
| chr11_118496296_118496447 | 0.14 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
10982 |
0.14 |
| chr7_24935298_24935521 | 0.14 |
Erfl |
ETS repressor factor like |
9085 |
0.09 |
| chr12_98562134_98562933 | 0.14 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
12179 |
0.14 |
| chr4_22485088_22485284 | 0.14 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
3180 |
0.21 |
| chr4_134470669_134471383 | 0.14 |
Stmn1 |
stathmin 1 |
1284 |
0.28 |
| chr10_93883117_93883275 | 0.14 |
Metap2 |
methionine aminopeptidase 2 |
4290 |
0.15 |
| chr6_56017510_56018185 | 0.14 |
Ppp1r17 |
protein phosphatase 1, regulatory subunit 17 |
350 |
0.93 |
| chr4_44247087_44247879 | 0.14 |
Rnf38 |
ring finger protein 38 |
13694 |
0.17 |
| chr17_25017468_25017619 | 0.14 |
Ift140 |
intraflagellar transport 140 |
1144 |
0.33 |
| chr3_118432535_118432944 | 0.14 |
Gm26871 |
predicted gene, 26871 |
1058 |
0.3 |
| chr6_129352639_129352989 | 0.14 |
Clec12a |
C-type lectin domain family 12, member a |
2850 |
0.17 |
| chr3_149190517_149191107 | 0.14 |
Gm42647 |
predicted gene 42647 |
19459 |
0.21 |
| chr17_93555961_93556112 | 0.14 |
Gm50002 |
predicted gene, 50002 |
124629 |
0.05 |
| chr2_93278081_93278912 | 0.14 |
Tspan18 |
tetraspanin 18 |
34019 |
0.17 |
| chr8_39514538_39514692 | 0.14 |
Gm5609 |
predicted gene 5609 |
58705 |
0.15 |
| chr11_32199697_32199897 | 0.14 |
Il9r |
interleukin 9 receptor |
425 |
0.73 |
| chr17_57769126_57770140 | 0.14 |
Cntnap5c |
contactin associated protein-like 5C |
63 |
0.97 |
| chr2_126490465_126490739 | 0.14 |
Atp8b4 |
ATPase, class I, type 8B, member 4 |
951 |
0.64 |
| chr16_64855062_64855213 | 0.14 |
Cggbp1 |
CGG triplet repeat binding protein 1 |
2435 |
0.26 |
| chr17_50967883_50968034 | 0.14 |
Tbc1d5 |
TBC1 domain family, member 5 |
16668 |
0.25 |
| chr15_103016875_103017501 | 0.14 |
Hoxc4 |
homeobox C4 |
1746 |
0.16 |
| chr4_22481988_22482455 | 0.13 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
6145 |
0.18 |
| chr2_61808611_61809537 | 0.13 |
Tbr1 |
T-box brain gene 1 |
2801 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.3 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.0 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |