Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
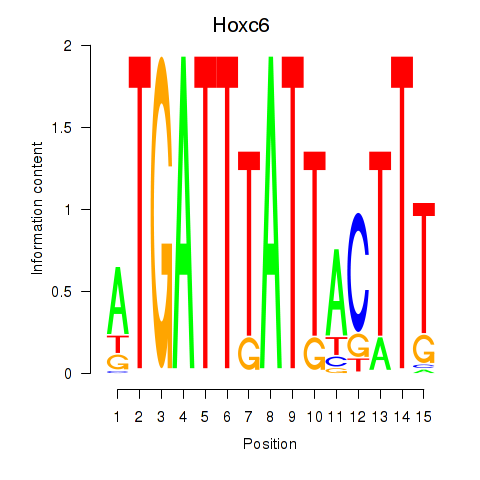
Results for Hoxc6
Z-value: 0.75
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.4 | homeobox C6 |
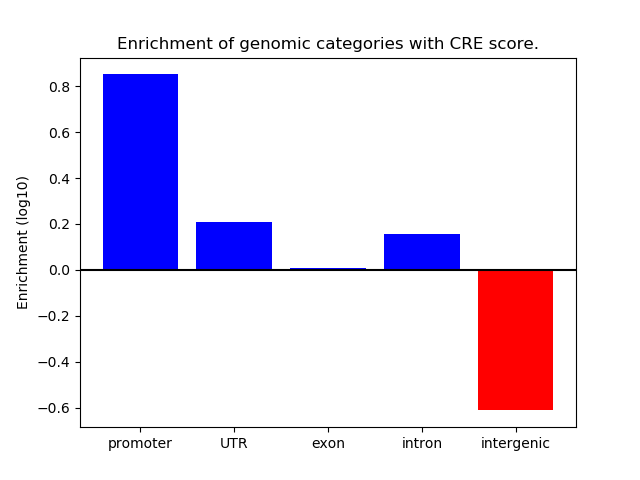
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_103003313_103003464 | Hoxc6 | 2859 | 0.114275 | -0.15 | 2.6e-01 | Click! |
| chr15_103009345_103011570 | Hoxc6 | 884 | 0.348197 | 0.07 | 5.9e-01 | Click! |
| chr15_103001653_103001804 | Hoxc6 | 1199 | 0.253116 | -0.07 | 6.0e-01 | Click! |
| chr15_102998770_103001153 | Hoxc6 | 568 | 0.536896 | -0.07 | 6.2e-01 | Click! |
| chr15_103001984_103002537 | Hoxc6 | 1731 | 0.171571 | -0.06 | 6.3e-01 | Click! |
Activity of the Hoxc6 motif across conditions
Conditions sorted by the z-value of the Hoxc6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
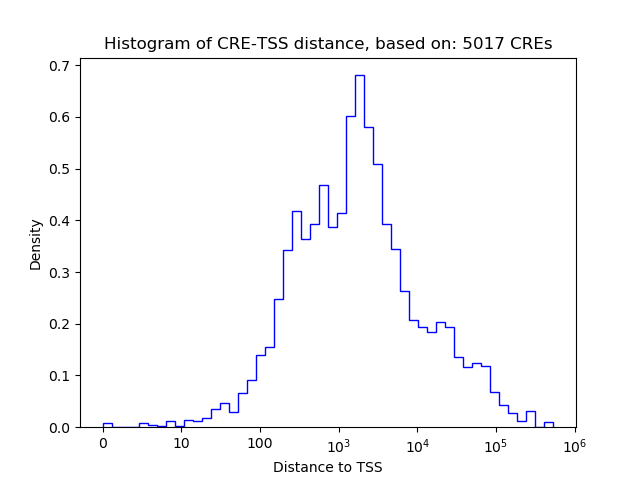
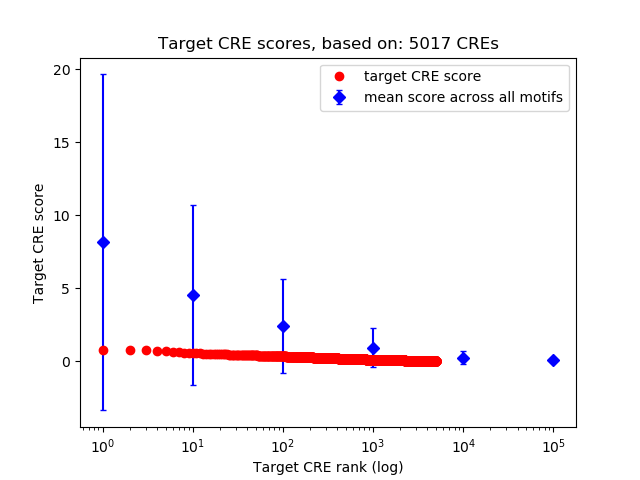
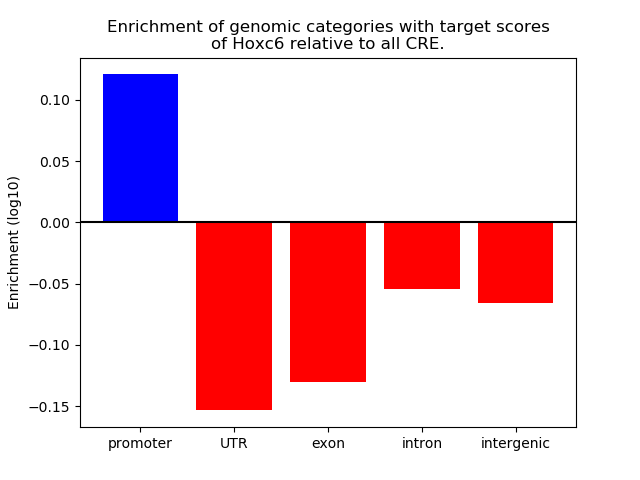
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_110753664_110754027 | 0.77 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
1304 |
0.42 |
| chr17_33953373_33953736 | 0.76 |
Rps18 |
ribosomal protein S18 |
865 |
0.21 |
| chr2_47228816_47228967 | 0.74 |
Gm25959 |
predicted gene, 25959 |
313093 |
0.01 |
| chr6_83532474_83532625 | 0.67 |
Actg2 |
actin, gamma 2, smooth muscle, enteric |
3674 |
0.14 |
| chr1_65188302_65188453 | 0.67 |
Pikfyve |
phosphoinositide kinase, FYVE type zinc finger containing |
1627 |
0.28 |
| chr10_55187657_55187808 | 0.66 |
Gm46190 |
predicted gene, 46190 |
63370 |
0.15 |
| chr17_24527137_24527295 | 0.60 |
Traf7 |
TNF receptor-associated factor 7 |
273 |
0.68 |
| chr2_144269311_144269468 | 0.57 |
Snx5 |
sorting nexin 5 |
1183 |
0.24 |
| chr6_65144371_65145258 | 0.56 |
Hpgds |
hematopoietic prostaglandin D synthase |
94 |
0.95 |
| chr8_80054187_80054695 | 0.55 |
Hhip |
Hedgehog-interacting protein |
3565 |
0.26 |
| chr15_103212708_103212859 | 0.54 |
Cbx5 |
chromobox 5 |
2522 |
0.18 |
| chr10_34296054_34296228 | 0.54 |
Tspyl4 |
TSPY-like 4 |
1301 |
0.27 |
| chr2_118972455_118972646 | 0.52 |
Gm14089 |
predicted gene 14089 |
24518 |
0.11 |
| chr3_25688317_25688468 | 0.52 |
Gm37573 |
predicted gene, 37573 |
11911 |
0.26 |
| chr19_53824390_53825419 | 0.51 |
Gm16299 |
predicted gene 16299 |
224 |
0.92 |
| chr2_103479284_103479435 | 0.50 |
Cat |
catalase |
5766 |
0.2 |
| chr5_67423933_67424105 | 0.50 |
Bend4 |
BEN domain containing 4 |
3311 |
0.16 |
| chr9_63613079_63613406 | 0.49 |
Aagab |
alpha- and gamma-adaptin binding protein |
3746 |
0.23 |
| chr16_65817251_65817521 | 0.49 |
Vgll3 |
vestigial like family member 3 |
1757 |
0.42 |
| chr10_102513522_102514467 | 0.48 |
Rassf9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
1738 |
0.36 |
| chr4_46453015_46453166 | 0.48 |
Anp32b |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
2188 |
0.22 |
| chr17_33891016_33891281 | 0.48 |
Kifc1 |
kinesin family member C1 |
487 |
0.48 |
| chr4_28818810_28818964 | 0.47 |
Epha7 |
Eph receptor A7 |
3458 |
0.27 |
| chr5_107836773_107836924 | 0.47 |
Evi5 |
ecotropic viral integration site 5 |
632 |
0.57 |
| chr4_88718352_88718503 | 0.46 |
Gm26566 |
predicted gene, 26566 |
3378 |
0.06 |
| chr17_35590766_35590985 | 0.46 |
Sfta2 |
surfactant associated 2 |
10674 |
0.08 |
| chr6_117880035_117880414 | 0.46 |
Zfp239 |
zinc finger protein 239 |
17140 |
0.1 |
| chr19_10201682_10201944 | 0.45 |
Fen1 |
flap structure specific endonuclease 1 |
2081 |
0.15 |
| chr10_78393243_78393558 | 0.44 |
Gm10146 |
predicted gene 10146 |
80 |
0.93 |
| chr16_81203532_81203683 | 0.44 |
Ncam2 |
neural cell adhesion molecule 2 |
2850 |
0.36 |
| chr17_34950362_34951201 | 0.44 |
Snord52 |
small nucleolar RNA, C/D box 52 |
236 |
0.58 |
| chr11_77987388_77987539 | 0.44 |
Phf12 |
PHD finger protein 12 |
4661 |
0.13 |
| chr14_93884726_93884877 | 0.44 |
Pcdh9 |
protocadherin 9 |
947 |
0.7 |
| chr11_48834664_48835112 | 0.44 |
Trim7 |
tripartite motif-containing 7 |
2577 |
0.13 |
| chr18_62927171_62927454 | 0.44 |
Apcdd1 |
adenomatosis polyposis coli down-regulated 1 |
4806 |
0.22 |
| chr2_50068247_50068603 | 0.43 |
Lypd6 |
LY6/PLAUR domain containing 6 |
1961 |
0.48 |
| chr4_152494393_152494544 | 0.43 |
Nphp4 |
nephronophthisis 4 (juvenile) homolog (human) |
6118 |
0.21 |
| chr3_20034383_20034767 | 0.43 |
Hps3 |
HPS3, biogenesis of lysosomal organelles complex 2 subunit 1 |
705 |
0.68 |
| chr1_24005709_24005937 | 0.42 |
Sdhaf4 |
succinate dehydrogenase complex assembly factor 4 |
167 |
0.68 |
| chr14_70894351_70894868 | 0.42 |
Gfra2 |
glial cell line derived neurotrophic factor family receptor alpha 2 |
4081 |
0.27 |
| chr9_85846725_85846928 | 0.42 |
9330154J02Rik |
RIKEN cDNA 9330154J02 gene |
2503 |
0.24 |
| chr10_55581070_55581221 | 0.41 |
Gm46190 |
predicted gene, 46190 |
456783 |
0.01 |
| chr8_88846133_88846460 | 0.41 |
Gm45504 |
predicted gene 45504 |
16778 |
0.21 |
| chr13_100650429_100650990 | 0.41 |
Ak6 |
adenylate kinase 6 |
270 |
0.46 |
| chr3_129724725_129724904 | 0.41 |
Egf |
epidermal growth factor |
263 |
0.88 |
| chr13_39979317_39979823 | 0.41 |
Gm47316 |
predicted gene, 47316 |
95189 |
0.08 |
| chr16_44357816_44357967 | 0.41 |
Spice1 |
spindle and centriole associated protein 1 |
9760 |
0.2 |
| chrX_6171274_6171425 | 0.40 |
Nudt10 |
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
1666 |
0.4 |
| chr7_37364441_37364642 | 0.40 |
6720469O03Rik |
RIKEN cDNA 6720469O03 gene |
2089 |
0.39 |
| chr4_58944015_58944496 | 0.40 |
Zkscan16 |
zinc finger with KRAB and SCAN domains 16 |
627 |
0.65 |
| chr13_119406449_119406600 | 0.40 |
Nnt |
nicotinamide nucleotide transhydrogenase |
2428 |
0.26 |
| chr2_26902153_26902850 | 0.40 |
Surf6 |
surfeit gene 6 |
289 |
0.76 |
| chr10_100589282_100589617 | 0.39 |
4930430F08Rik |
RIKEN cDNA 4930430F08 gene |
54 |
0.94 |
| chr15_32020772_32021024 | 0.39 |
Gm49285 |
predicted gene, 49285 |
29053 |
0.23 |
| chr13_21906553_21907020 | 0.39 |
Gm11279 |
predicted gene 11279 |
3630 |
0.08 |
| chr1_20762743_20762894 | 0.39 |
Il17f |
interleukin 17F |
16736 |
0.13 |
| chr16_76866379_76866591 | 0.39 |
1700041M19Rik |
RIKEN cDNA 1700041M19 gene |
22796 |
0.18 |
| chr6_115545196_115545422 | 0.39 |
Tsen2 |
tRNA splicing endonuclease subunit 2 |
603 |
0.67 |
| chr13_45849819_45850172 | 0.39 |
Atxn1 |
ataxin 1 |
22293 |
0.23 |
| chr4_91058550_91058701 | 0.38 |
Gm12643 |
predicted gene 12643 |
20725 |
0.23 |
| chr14_30063443_30063594 | 0.38 |
Cacna1d |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
7004 |
0.15 |
| chr5_17220718_17220869 | 0.38 |
Gm6673 |
predicted gene 6673 |
7141 |
0.31 |
| chr4_130665263_130665565 | 0.38 |
Pum1 |
pumilio RNA-binding family member 1 |
2008 |
0.35 |
| chr18_6514668_6515014 | 0.38 |
Epc1 |
enhancer of polycomb homolog 1 |
1267 |
0.42 |
| chr5_100568256_100568429 | 0.38 |
Plac8 |
placenta-specific 8 |
3850 |
0.16 |
| chr6_49368197_49368627 | 0.38 |
Fam221a |
family with sequence similarity 221, member A |
516 |
0.76 |
| chr11_88051697_88051848 | 0.37 |
Srsf1 |
serine and arginine-rich splicing factor 1 |
2360 |
0.19 |
| chr8_74877423_74877574 | 0.37 |
Isx |
intestine specific homeobox |
3958 |
0.28 |
| chr4_129237589_129237740 | 0.37 |
C77080 |
expressed sequence C77080 |
1510 |
0.28 |
| chr1_163929450_163929750 | 0.37 |
Scyl3 |
SCY1-like 3 (S. cerevisiae) |
165 |
0.96 |
| chr13_109927479_109928182 | 0.37 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
986 |
0.65 |
| chr6_30543844_30544457 | 0.37 |
Cpa2 |
carboxypeptidase A2, pancreatic |
234 |
0.89 |
| chr3_129201620_129202526 | 0.37 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
1874 |
0.31 |
| chr4_58550341_58551468 | 0.36 |
Lpar1 |
lysophosphatidic acid receptor 1 |
2280 |
0.31 |
| chr17_75551079_75551471 | 0.36 |
Fam98a |
family with sequence similarity 98, member A |
521 |
0.85 |
| chr16_38903808_38903959 | 0.36 |
Igsf11 |
immunoglobulin superfamily, member 11 |
1375 |
0.43 |
| chr7_139388305_139388676 | 0.36 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
619 |
0.79 |
| chr3_34485769_34486286 | 0.36 |
Gm29135 |
predicted gene 29135 |
3820 |
0.23 |
| chr8_126665301_126665615 | 0.36 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
71472 |
0.1 |
| chr11_6527621_6527833 | 0.36 |
Snhg15 |
small nucleolar RNA host gene 15 |
32 |
0.85 |
| chr4_119188779_119189142 | 0.36 |
Ermap |
erythroblast membrane-associated protein |
213 |
0.86 |
| chr19_12794332_12794483 | 0.35 |
Zfp91 |
zinc finger protein 91 |
1719 |
0.21 |
| chr10_67176618_67176769 | 0.35 |
Jmjd1c |
jumonji domain containing 1C |
9055 |
0.22 |
| chr9_47625241_47625392 | 0.35 |
Cadm1 |
cell adhesion molecule 1 |
94943 |
0.07 |
| chrX_49272778_49272929 | 0.35 |
Enox2 |
ecto-NOX disulfide-thiol exchanger 2 |
15359 |
0.24 |
| chr2_150786328_150786674 | 0.35 |
Pygb |
brain glycogen phosphorylase |
234 |
0.91 |
| chr4_48472485_48473051 | 0.35 |
Tex10 |
testis expressed gene 10 |
575 |
0.81 |
| chr1_63129559_63129868 | 0.35 |
Gm20342 |
predicted gene, 20342 |
12595 |
0.09 |
| chr19_7495247_7495398 | 0.34 |
Atl3 |
atlastin GTPase 3 |
787 |
0.56 |
| chr7_120981024_120981392 | 0.34 |
Cdr2 |
cerebellar degeneration-related 2 |
582 |
0.54 |
| chr4_43847679_43847830 | 0.34 |
Olfr155 |
olfactory receptor 155 |
3811 |
0.13 |
| chr19_4809602_4809914 | 0.34 |
Gm21992 |
predicted gene 21992 |
1454 |
0.18 |
| chrX_95658567_95659171 | 0.34 |
Zc4h2 |
zinc finger, C4H2 domain containing |
360 |
0.88 |
| chr5_23851483_23851729 | 0.34 |
Snord93 |
small nucleolar RNA, C/D box 93 |
604 |
0.45 |
| chr15_50882452_50882685 | 0.34 |
Trps1 |
transcriptional repressor GATA binding 1 |
238 |
0.93 |
| chr6_96113911_96115198 | 0.34 |
Tafa1 |
TAFA chemokine like family member 1 |
95 |
0.98 |
| chr18_36746468_36746623 | 0.34 |
Ik |
IK cytokine |
1785 |
0.14 |
| chr7_19675210_19675386 | 0.34 |
Apoc2 |
apolipoprotein C-II |
1378 |
0.2 |
| chr13_21710772_21711935 | 0.33 |
H2bc13 |
H2B clustered histone 13 |
4790 |
0.06 |
| chr8_124099096_124099358 | 0.33 |
Gm45732 |
predicted gene 45732 |
50815 |
0.09 |
| chr7_36710059_36710756 | 0.33 |
Gm37452 |
predicted gene, 37452 |
233 |
0.9 |
| chr3_104434194_104434423 | 0.33 |
Gm9273 |
predicted gene 9273 |
15981 |
0.12 |
| chrX_113304333_113304650 | 0.33 |
Dach2 |
dachshund family transcription factor 2 |
5857 |
0.24 |
| chr2_39066898_39067049 | 0.33 |
Golga1 |
golgi autoantigen, golgin subfamily a, 1 |
1432 |
0.31 |
| chr5_73337784_73337935 | 0.33 |
Ociad2 |
OCIA domain containing 2 |
699 |
0.55 |
| chr6_47814323_47814474 | 0.33 |
Pdia4 |
protein disulfide isomerase associated 4 |
968 |
0.33 |
| chr2_4562127_4563316 | 0.33 |
Frmd4a |
FERM domain containing 4A |
2946 |
0.28 |
| chr1_171035524_171035675 | 0.33 |
Gm26110 |
predicted gene, 26110 |
7634 |
0.1 |
| chr19_60787666_60787822 | 0.32 |
Eif3a |
eukaryotic translation initiation factor 3, subunit A |
2899 |
0.15 |
| chr7_3285392_3285556 | 0.32 |
Gm5340 |
predicted gene 5340 |
0 |
0.94 |
| chrX_94212994_94213232 | 0.32 |
Eif2s3x |
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
251 |
0.91 |
| chr5_77310564_77310969 | 0.32 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
275 |
0.74 |
| chr11_71031860_71032011 | 0.32 |
6330403K07Rik |
RIKEN cDNA 6330403K07 gene |
1578 |
0.22 |
| chr4_109003238_109003389 | 0.32 |
Nrd1 |
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
2543 |
0.2 |
| chr2_156617951_156618102 | 0.32 |
Gm14174 |
predicted gene 14174 |
2578 |
0.17 |
| chr8_60957853_60958004 | 0.32 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
2695 |
0.23 |
| chr17_45725964_45726475 | 0.32 |
1600014C23Rik |
RIKEN cDNA 1600014C23 gene |
7625 |
0.13 |
| chr3_14864298_14864449 | 0.32 |
Car3 |
carbonic anhydrase 3 |
835 |
0.62 |
| chr17_84083532_84083802 | 0.32 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
5007 |
0.17 |
| chr11_19760689_19760940 | 0.32 |
Gm12028 |
predicted gene 12028 |
10748 |
0.19 |
| chr6_145360341_145360492 | 0.32 |
Gm23498 |
predicted gene, 23498 |
6442 |
0.15 |
| chr7_70354508_70355670 | 0.32 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
148 |
0.93 |
| chr8_40187126_40187473 | 0.32 |
Fgf20 |
fibroblast growth factor 20 |
99654 |
0.07 |
| chr10_100050584_100050771 | 0.32 |
Kitl |
kit ligand |
34761 |
0.14 |
| chr13_6549153_6549438 | 0.31 |
Pitrm1 |
pitrilysin metallepetidase 1 |
1060 |
0.54 |
| chr14_60250603_60250845 | 0.31 |
Gm49336 |
predicted gene, 49336 |
682 |
0.4 |
| chr2_34368049_34368262 | 0.31 |
Pbx3 |
pre B cell leukemia homeobox 3 |
2691 |
0.28 |
| chr8_111034573_111034976 | 0.31 |
Aars |
alanyl-tRNA synthetase |
858 |
0.4 |
| chr9_57520819_57521116 | 0.31 |
Cox5a |
cytochrome c oxidase subunit 5A |
307 |
0.79 |
| chr16_77597699_77598173 | 0.31 |
Mir99a |
microRNA 99a |
1000 |
0.31 |
| chr11_23633785_23634057 | 0.31 |
0610010F05Rik |
RIKEN cDNA 0610010F05 gene |
282 |
0.88 |
| chr11_85640273_85640424 | 0.30 |
Bcas3 |
breast carcinoma amplified sequence 3 |
356 |
0.9 |
| chr5_90519923_90520403 | 0.30 |
Afm |
afamin |
1214 |
0.35 |
| chr13_49609766_49610088 | 0.30 |
Ogn |
osteoglycin |
1881 |
0.26 |
| chr13_21788176_21788327 | 0.30 |
Hist1h2bp |
histone cluster 1, H2bp |
743 |
0.24 |
| chr5_87532219_87532460 | 0.30 |
Sult1b1 |
sulfotransferase family 1B, member 1 |
2807 |
0.17 |
| chr6_87848769_87849239 | 0.30 |
Cnbp |
cellular nucleic acid binding protein |
2018 |
0.14 |
| chr16_94372818_94373028 | 0.30 |
Pigp |
phosphatidylinositol glycan anchor biosynthesis, class P |
1081 |
0.39 |
| chr1_183389670_183389910 | 0.30 |
Taf1a |
TATA-box binding protein associated factor, RNA polymerase I, A |
805 |
0.52 |
| chr5_129847745_129847896 | 0.30 |
Sumf2 |
sulfatase modifying factor 2 |
800 |
0.44 |
| chr5_151367699_151368449 | 0.30 |
1700028E10Rik |
RIKEN cDNA 1700028E10 gene |
601 |
0.61 |
| chr15_50886808_50887208 | 0.30 |
Trps1 |
transcriptional repressor GATA binding 1 |
2038 |
0.29 |
| chr3_49840659_49840810 | 0.30 |
Gm37550 |
predicted gene, 37550 |
59847 |
0.13 |
| chr2_15056734_15057204 | 0.30 |
Gm38257 |
predicted gene, 38257 |
256 |
0.85 |
| chr10_39162766_39163414 | 0.30 |
Ccn6 |
cellular communication network factor 6 |
704 |
0.6 |
| chr2_172824721_172824976 | 0.30 |
Gm22773 |
predicted gene, 22773 |
39493 |
0.18 |
| chr13_100783304_100783455 | 0.30 |
Ccnb1 |
cyclin B1 |
1346 |
0.3 |
| chr19_14595420_14595901 | 0.30 |
Tle4 |
transducin-like enhancer of split 4 |
121 |
0.98 |
| chr18_6238794_6238945 | 0.30 |
Gm41664 |
predicted gene, 41664 |
1945 |
0.28 |
| chr15_82244872_82245028 | 0.30 |
Cenpm |
centromere protein M |
202 |
0.86 |
| chr7_29237918_29238107 | 0.29 |
Yif1b |
Yip1 interacting factor homolog B (S. cerevisiae) |
317 |
0.77 |
| chr10_62798201_62798352 | 0.29 |
Ccar1 |
cell division cycle and apoptosis regulator 1 |
5990 |
0.12 |
| chr12_92303420_92303633 | 0.29 |
Gm6841 |
predicted gene 6841 |
84149 |
0.1 |
| chr15_80254231_80254977 | 0.29 |
Atf4 |
activating transcription factor 4 |
580 |
0.6 |
| chr7_15946126_15946297 | 0.29 |
Nop53 |
NOP53 ribosome biogenesis factor |
137 |
0.92 |
| chr10_15300720_15300871 | 0.29 |
Gm47710 |
predicted gene, 47710 |
398 |
0.91 |
| chr5_66077391_66078254 | 0.29 |
Rbm47 |
RNA binding motif protein 47 |
3162 |
0.16 |
| chr11_77511266_77511543 | 0.29 |
Trp53i13 |
transformation related protein 53 inducible protein 13 |
1359 |
0.27 |
| chr12_36315313_36316308 | 0.29 |
Sostdc1 |
sclerostin domain containing 1 |
1671 |
0.31 |
| chr18_49831854_49832487 | 0.29 |
Dmxl1 |
Dmx-like 1 |
500 |
0.82 |
| chr10_119645936_119646087 | 0.29 |
Gm47026 |
predicted gene, 47026 |
8053 |
0.2 |
| chr1_171057110_171057261 | 0.29 |
Fcgr3 |
Fc receptor, IgG, low affinity III |
869 |
0.4 |
| chr18_31633443_31633594 | 0.29 |
Sap130 |
Sin3A associated protein |
853 |
0.58 |
| chr11_58014866_58015017 | 0.29 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
5877 |
0.17 |
| chrX_103621531_103622429 | 0.29 |
Ftx |
Ftx transcript, Xist regulator (non-protein coding) |
389 |
0.72 |
| chr7_4813003_4813336 | 0.29 |
Ube2s |
ubiquitin-conjugating enzyme E2S |
579 |
0.51 |
| chr6_73746297_73746478 | 0.29 |
Gm31747 |
predicted gene, 31747 |
137714 |
0.05 |
| chr2_121474102_121474346 | 0.28 |
Mfap1b |
microfibrillar-associated protein 1B |
157 |
0.88 |
| chr2_109283503_109283700 | 0.28 |
Kif18a |
kinesin family member 18A |
2732 |
0.29 |
| chr13_41219553_41219704 | 0.28 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
534 |
0.68 |
| chr6_86815190_86815697 | 0.28 |
Anxa4 |
annexin A4 |
21859 |
0.12 |
| chr4_40301017_40301186 | 0.28 |
4930509K18Rik |
RIKEN cDNA 4930509K18 gene |
12131 |
0.15 |
| chr8_33652198_33652349 | 0.28 |
Gsr |
glutathione reductase |
250 |
0.9 |
| chr14_19709512_19709663 | 0.28 |
Gm49341 |
predicted gene, 49341 |
12378 |
0.13 |
| chr1_125436417_125436590 | 0.28 |
Actr3 |
ARP3 actin-related protein 3 |
776 |
0.71 |
| chrX_65691240_65691391 | 0.28 |
Gm14668 |
predicted gene 14668 |
245034 |
0.02 |
| chr9_73039777_73040017 | 0.28 |
Pigbos1 |
Pigb opposite strand 1 |
167 |
0.52 |
| chr8_79829805_79829956 | 0.28 |
Gm45433 |
predicted gene 45433 |
26269 |
0.14 |
| chr17_53481120_53481271 | 0.28 |
Rab5a |
RAB5A, member RAS oncogene family |
1908 |
0.25 |
| chr1_180365356_180365966 | 0.28 |
Gm37390 |
predicted gene, 37390 |
7008 |
0.13 |
| chr9_19804902_19805053 | 0.28 |
Olfr859 |
olfactory receptor 859 |
69 |
0.96 |
| chr9_44113725_44113909 | 0.28 |
Rnf26 |
ring finger protein 26 |
300 |
0.41 |
| chr15_50891999_50892241 | 0.27 |
Trps1 |
transcriptional repressor GATA binding 1 |
1657 |
0.36 |
| chr11_108621492_108621702 | 0.27 |
Cep112 |
centrosomal protein 112 |
15818 |
0.22 |
| chr10_4347012_4347226 | 0.27 |
Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
311 |
0.88 |
| chr17_33393976_33394596 | 0.27 |
Zfp101 |
zinc finger protein 101 |
71 |
0.97 |
| chr9_116903598_116903749 | 0.27 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
80854 |
0.11 |
| chr9_71976137_71976437 | 0.27 |
Gm37663 |
predicted gene, 37663 |
14984 |
0.1 |
| chr12_87443920_87444600 | 0.27 |
Alkbh1 |
alkB homolog 1, histone H2A dioxygenase |
243 |
0.52 |
| chr5_130256689_130256849 | 0.27 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
266 |
0.82 |
| chr1_19219527_19219822 | 0.27 |
Tfap2b |
transcription factor AP-2 beta |
5795 |
0.21 |
| chr8_12671228_12671386 | 0.27 |
Tubgcp3 |
tubulin, gamma complex associated protein 3 |
652 |
0.48 |
| chr16_10447447_10447926 | 0.27 |
Tvp23a |
trans-golgi network vesicle protein 23A |
324 |
0.87 |
| chr2_61514942_61515095 | 0.27 |
Gm22338 |
predicted gene, 22338 |
22252 |
0.23 |
| chr11_101626644_101627106 | 0.27 |
Rdm1 |
RAD52 motif 1 |
320 |
0.56 |
| chr11_49246951_49247357 | 0.27 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
323 |
0.83 |
| chr1_38624448_38624617 | 0.27 |
Aff3 |
AF4/FMR2 family, member 3 |
2653 |
0.36 |
| chr12_5376777_5377685 | 0.27 |
2810032G03Rik |
RIKEN cDNA 2810032G03 gene |
1361 |
0.39 |
| chr5_123748067_123748318 | 0.27 |
Rsrc2 |
arginine/serine-rich coiled-coil 2 |
1175 |
0.32 |
| chr17_46160338_46160791 | 0.27 |
Gtpbp2 |
GTP binding protein 2 |
468 |
0.68 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.3 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.3 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.1 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.2 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.2 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.0 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.2 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0001802 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.0 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.0 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0032762 | mast cell cytokine production(GO:0032762) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.4 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.2 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0045713 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.0 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:1902805 | positive regulation of synaptic vesicle transport(GO:1902805) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.0 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.0 | 0.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.0 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.0 | 0.0 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.0 | GO:0061083 | regulation of protein refolding(GO:0061083) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.0 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 0.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0044466 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0051731 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0032357 | oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.0 | 0.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |