Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
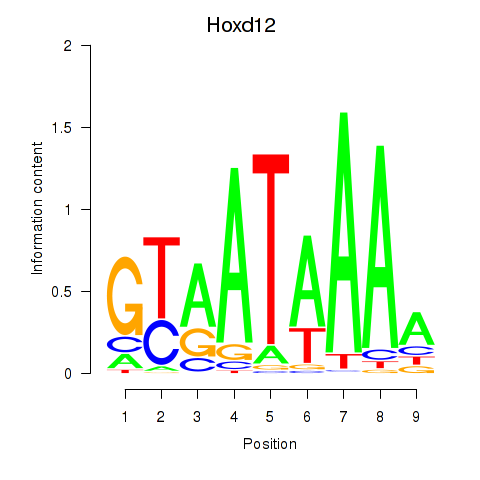
Results for Hoxd12
Z-value: 0.50
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd12
|
ENSMUSG00000001823.4 | homeobox D12 |
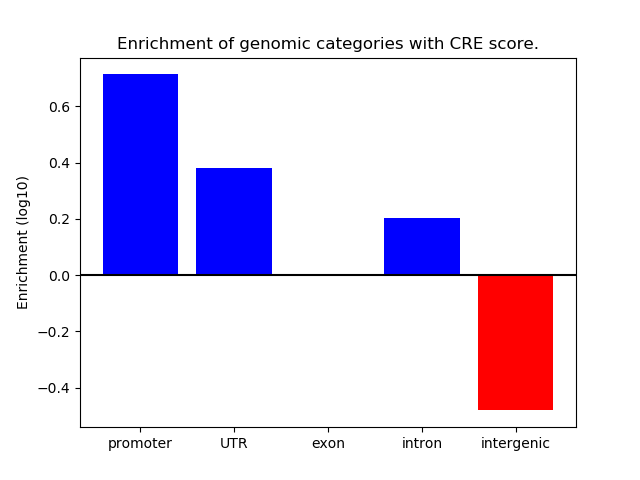
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_74673537_74673688 | Hoxd12 | 1401 | 0.158620 | 0.26 | 4.5e-02 | Click! |
| chr2_74672425_74673160 | Hoxd12 | 2221 | 0.099583 | 0.24 | 6.3e-02 | Click! |
| chr2_74673325_74673476 | Hoxd12 | 1613 | 0.135621 | 0.24 | 6.6e-02 | Click! |
| chr2_74676368_74676604 | Hoxd12 | 1473 | 0.145217 | 0.21 | 1.1e-01 | Click! |
| chr2_74675065_74676326 | Hoxd12 | 682 | 0.366054 | 0.20 | 1.3e-01 | Click! |
Activity of the Hoxd12 motif across conditions
Conditions sorted by the z-value of the Hoxd12 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
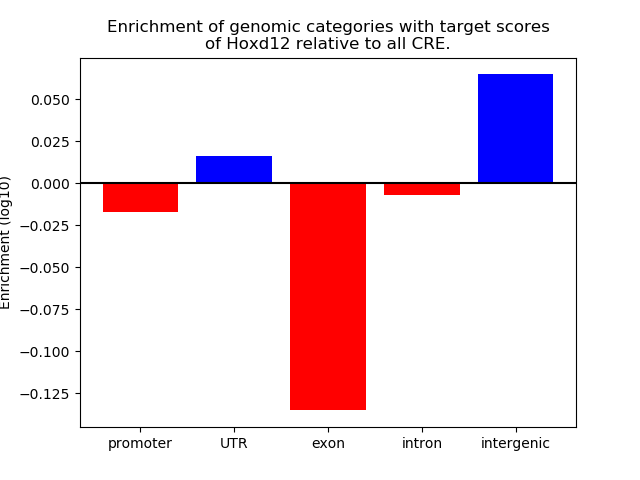
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_21485587_21486692 | 0.82 |
Agtr2 |
angiotensin II receptor, type 2 |
1417 |
0.41 |
| chr6_16797640_16797838 | 0.63 |
Gm36669 |
predicted gene, 36669 |
20215 |
0.22 |
| chr10_20723138_20723899 | 0.60 |
Pde7b |
phosphodiesterase 7B |
1178 |
0.55 |
| chr18_49117603_49118121 | 0.55 |
Gm18993 |
predicted gene, 18993 |
54438 |
0.18 |
| chr2_74063525_74063783 | 0.55 |
A630050E04Rik |
RIKEN cDNA A630050E04 gene |
38688 |
0.15 |
| chr5_3432895_3433605 | 0.51 |
Cdk6 |
cyclin-dependent kinase 6 |
37699 |
0.12 |
| chr3_48846231_48846444 | 0.50 |
Gm37190 |
predicted gene, 37190 |
85388 |
0.1 |
| chr16_74408614_74409150 | 0.50 |
Robo2 |
roundabout guidance receptor 2 |
2030 |
0.43 |
| chr12_36315313_36316308 | 0.48 |
Sostdc1 |
sclerostin domain containing 1 |
1671 |
0.31 |
| chr5_48003784_48004411 | 0.48 |
4930518C09Rik |
RIKEN cDNA 4930518C09 gene |
1610 |
0.28 |
| chr8_85698168_85698436 | 0.46 |
Neto2 |
neuropilin (NRP) and tolloid (TLL)-like 2 |
2452 |
0.21 |
| chr18_40258245_40258964 | 0.45 |
Gm31087 |
predicted gene, 31087 |
109 |
0.63 |
| chr10_125967599_125967789 | 0.44 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1441 |
0.55 |
| chrX_104198423_104198882 | 0.44 |
Nexmif |
neurite extension and migration factor |
2465 |
0.34 |
| chr4_84403786_84404211 | 0.43 |
Gm12421 |
predicted gene 12421 |
25126 |
0.25 |
| chr8_65036064_65036521 | 0.40 |
Gm45886 |
predicted gene 45886 |
804 |
0.33 |
| chr18_60377377_60377703 | 0.40 |
Iigp1 |
interferon inducible GTPase 1 |
1493 |
0.33 |
| chr3_149705320_149706167 | 0.40 |
Gm31121 |
predicted gene, 31121 |
45347 |
0.2 |
| chr5_32059174_32059703 | 0.40 |
Babam2 |
BRISC and BRCA1 A complex member 2 |
13460 |
0.19 |
| chr16_77593272_77593433 | 0.40 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
476 |
0.66 |
| chr1_168425173_168425741 | 0.40 |
Pbx1 |
pre B cell leukemia homeobox 1 |
6047 |
0.27 |
| chr2_169099862_169100013 | 0.39 |
Gm14258 |
predicted gene 14258 |
6516 |
0.23 |
| chr1_143640264_143641520 | 0.39 |
B3galt2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
228 |
0.59 |
| chr6_49009229_49009402 | 0.39 |
4930563H07Rik |
RIKEN cDNA 4930563H07 gene |
9699 |
0.08 |
| chr1_21500023_21500174 | 0.38 |
Gm7658 |
predicted gene 7658 |
12958 |
0.19 |
| chr14_108415348_108415499 | 0.38 |
Gm24818 |
predicted gene, 24818 |
35859 |
0.24 |
| chr17_51461105_51461697 | 0.37 |
Gm46574 |
predicted gene, 46574 |
23320 |
0.19 |
| chr10_59950135_59950286 | 0.36 |
Ddit4 |
DNA-damage-inducible transcript 4 |
1624 |
0.36 |
| chr16_92692721_92692872 | 0.36 |
Runx1 |
runt related transcription factor 1 |
2699 |
0.35 |
| chr8_91948462_91949309 | 0.36 |
Gm28515 |
predicted gene 28515 |
35819 |
0.12 |
| chr8_47351619_47352381 | 0.36 |
Stox2 |
storkhead box 2 |
348 |
0.91 |
| chr4_134372795_134372981 | 0.35 |
Extl1 |
exostosin-like glycosyltransferase 1 |
330 |
0.82 |
| chr6_6880959_6882181 | 0.35 |
Dlx5 |
distal-less homeobox 5 |
498 |
0.71 |
| chr7_118902757_118903362 | 0.34 |
Iqck |
IQ motif containing K |
34064 |
0.13 |
| chr14_80901423_80902185 | 0.34 |
Gm49044 |
predicted gene, 49044 |
28933 |
0.21 |
| chr4_81566839_81567150 | 0.33 |
Gm11765 |
predicted gene 11765 |
105262 |
0.07 |
| chr3_149443059_149443897 | 0.33 |
Gm30382 |
predicted gene, 30382 |
1798 |
0.48 |
| chr5_44796139_44796769 | 0.33 |
Ldb2 |
LIM domain binding 2 |
3148 |
0.24 |
| chr19_20009817_20010437 | 0.32 |
Gm22684 |
predicted gene, 22684 |
23508 |
0.22 |
| chr8_58909555_58909706 | 0.32 |
Galntl6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
1946 |
0.34 |
| chr3_149358058_149358806 | 0.32 |
Gm26468 |
predicted gene, 26468 |
17424 |
0.19 |
| chr15_21110687_21111815 | 0.32 |
Cdh12 |
cadherin 12 |
201 |
0.94 |
| chr3_86759901_86760193 | 0.32 |
Gm37876 |
predicted gene, 37876 |
18606 |
0.19 |
| chr12_34385779_34385930 | 0.32 |
Hdac9 |
histone deacetylase 9 |
51453 |
0.17 |
| chr5_119690061_119690212 | 0.32 |
Tbx3os2 |
T-box 3, opposite strand 2 |
1082 |
0.45 |
| chr16_74070800_74070951 | 0.32 |
Gm49660 |
predicted gene, 49660 |
5022 |
0.22 |
| chr19_14670192_14670662 | 0.32 |
Gm37997 |
predicted gene, 37997 |
11760 |
0.25 |
| chr1_186546580_186547168 | 0.32 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
11598 |
0.25 |
| chr6_129450832_129451818 | 0.32 |
Clec1a |
C-type lectin domain family 1, member a |
510 |
0.65 |
| chrX_88113433_88114223 | 0.31 |
Il1rapl1 |
interleukin 1 receptor accessory protein-like 1 |
1817 |
0.46 |
| chr11_86472817_86473930 | 0.31 |
Rnft1 |
ring finger protein, transmembrane 1 |
11284 |
0.16 |
| chr5_18358524_18358675 | 0.31 |
Gnai1 |
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
1756 |
0.43 |
| chr15_43662128_43662279 | 0.31 |
Gm2140 |
predicted gene 2140 |
146258 |
0.04 |
| chr2_46442289_46442440 | 0.31 |
Gm13470 |
predicted gene 13470 |
346 |
0.92 |
| chr2_137356841_137357676 | 0.31 |
Gm14055 |
predicted gene 14055 |
68755 |
0.13 |
| chr18_37481682_37481833 | 0.31 |
Pcdhb17 |
protocadherin beta 17 |
3038 |
0.09 |
| chr11_24980356_24980734 | 0.31 |
Gm12665 |
predicted gene 12665 |
54487 |
0.17 |
| chr15_37231016_37231198 | 0.30 |
Grhl2 |
grainyhead like transcription factor 2 |
1929 |
0.24 |
| chr15_39391612_39392273 | 0.30 |
Rims2 |
regulating synaptic membrane exocytosis 2 |
142 |
0.97 |
| chr15_102594215_102594505 | 0.29 |
Atf7 |
activating transcription factor 7 |
30562 |
0.08 |
| chr18_83557517_83557756 | 0.29 |
Gm50416 |
predicted gene, 50416 |
32332 |
0.14 |
| chr1_69591259_69591484 | 0.29 |
Gm29112 |
predicted gene 29112 |
7151 |
0.24 |
| chr1_69790500_69791180 | 0.29 |
Gm37930 |
predicted gene, 37930 |
19972 |
0.21 |
| chr17_85693787_85694529 | 0.28 |
CJ186046Rik |
Riken cDNA CJ186046 gene |
529 |
0.77 |
| chr1_119052514_119053221 | 0.28 |
Gli2 |
GLI-Kruppel family member GLI2 |
472 |
0.83 |
| chr3_42074735_42075052 | 0.28 |
Gm37846 |
predicted gene, 37846 |
88972 |
0.08 |
| chr16_65560252_65560595 | 0.28 |
Chmp2b |
charged multivesicular body protein 2B |
2097 |
0.32 |
| chr18_69349288_69349439 | 0.28 |
Tcf4 |
transcription factor 4 |
419 |
0.89 |
| chr17_64329718_64329909 | 0.28 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
2052 |
0.38 |
| chr15_81936444_81938042 | 0.28 |
Csdc2 |
cold shock domain containing C2, RNA binding |
261 |
0.82 |
| chr2_44558382_44559022 | 0.28 |
Gtdc1 |
glycosyltransferase-like domain containing 1 |
12645 |
0.3 |
| chr16_89958503_89958654 | 0.28 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
2159 |
0.43 |
| chr12_47373270_47373421 | 0.28 |
Gm48578 |
predicted gene, 48578 |
158912 |
0.04 |
| chr1_14357767_14358207 | 0.27 |
Eya1 |
EYA transcriptional coactivator and phosphatase 1 |
47752 |
0.18 |
| chr14_36918744_36919600 | 0.27 |
Ccser2 |
coiled-coil serine rich 2 |
161 |
0.97 |
| chr17_43571796_43572049 | 0.27 |
Pla2g7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
3207 |
0.21 |
| chr18_32941213_32941434 | 0.27 |
2310026I22Rik |
RIKEN cDNA 2310026I22 gene |
2094 |
0.27 |
| chr9_103212682_103213306 | 0.27 |
Trf |
transferrin |
988 |
0.51 |
| chr2_59438620_59439113 | 0.27 |
Gm13549 |
predicted gene 13549 |
39085 |
0.13 |
| chr17_3397051_3398448 | 0.27 |
Tiam2 |
T cell lymphoma invasion and metastasis 2 |
519 |
0.8 |
| chr13_49610100_49610769 | 0.27 |
Ogn |
osteoglycin |
2388 |
0.21 |
| chr9_66004589_66004841 | 0.27 |
Csnk1g1 |
casein kinase 1, gamma 1 |
3025 |
0.23 |
| chr14_7774273_7774663 | 0.26 |
Gm10044 |
predicted gene 10044 |
329 |
0.85 |
| chr12_101815999_101816150 | 0.26 |
Fbln5 |
fibulin 5 |
2394 |
0.3 |
| chr3_63851016_63851674 | 0.26 |
Plch1 |
phospholipase C, eta 1 |
14 |
0.97 |
| chr2_135661117_135661413 | 0.26 |
Plcb4 |
phospholipase C, beta 4 |
1554 |
0.35 |
| chr7_115920406_115920948 | 0.26 |
Sox6 |
SRY (sex determining region Y)-box 6 |
60825 |
0.12 |
| chr18_57785762_57786717 | 0.26 |
Gm19519 |
predicted gene, 19519 |
6094 |
0.27 |
| chr7_16948879_16949030 | 0.26 |
Pnmal2 |
PNMA-like 2 |
4272 |
0.1 |
| chr5_49284738_49286021 | 0.26 |
Kcnip4 |
Kv channel interacting protein 4 |
280 |
0.92 |
| chr5_116312100_116312726 | 0.26 |
B230112J18Rik |
RIKEN cDNA B230112J18 gene |
53 |
0.96 |
| chr5_76823713_76823864 | 0.26 |
C530008M17Rik |
RIKEN cDNA C530008M17 gene |
13800 |
0.19 |
| chr7_45627553_45628284 | 0.26 |
Rasip1 |
Ras interacting protein 1 |
430 |
0.56 |
| chr15_16730998_16731546 | 0.25 |
Cdh9 |
cadherin 9 |
2516 |
0.41 |
| chr10_42861990_42862751 | 0.25 |
Scml4 |
Scm polycomb group protein like 4 |
1577 |
0.26 |
| chr8_50913867_50914080 | 0.25 |
C130073E24Rik |
RIKEN cDNA C130073E24 gene |
3024 |
0.4 |
| chr5_150260534_150260992 | 0.25 |
Fry |
FRY microtubule binding protein |
996 |
0.55 |
| chr13_91916211_91916382 | 0.25 |
Ckmt2 |
creatine kinase, mitochondrial 2 |
39411 |
0.15 |
| chr8_94265837_94266069 | 0.25 |
Nup93 |
nucleoporin 93 |
894 |
0.46 |
| chr14_101841178_101841822 | 0.25 |
Lmo7 |
LIM domain only 7 |
681 |
0.79 |
| chr2_27890316_27890530 | 0.25 |
2810430I11Rik |
RIKEN cDNA 2810430I11 gene |
3371 |
0.26 |
| chr7_69845047_69845447 | 0.25 |
Gm44690 |
predicted gene 44690 |
39113 |
0.19 |
| chr4_76440840_76441299 | 0.25 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
8911 |
0.22 |
| chr13_97216191_97216342 | 0.25 |
Gm48881 |
predicted gene, 48881 |
13291 |
0.14 |
| chr3_116923152_116923435 | 0.25 |
Gm42892 |
predicted gene 42892 |
21037 |
0.12 |
| chr2_16358886_16359826 | 0.25 |
Plxdc2 |
plexin domain containing 2 |
2239 |
0.48 |
| chr1_17601856_17602960 | 0.25 |
Pi15 |
peptidase inhibitor 15 |
507 |
0.82 |
| chr12_95699008_95699374 | 0.24 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
3834 |
0.21 |
| chr16_93272287_93272901 | 0.24 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
80595 |
0.09 |
| chr3_149257135_149257363 | 0.24 |
Gm10287 |
predicted gene 10287 |
31504 |
0.16 |
| chr16_28751131_28751282 | 0.24 |
Fgf12 |
fibroblast growth factor 12 |
1862 |
0.48 |
| chr10_103020026_103020270 | 0.24 |
Alx1 |
ALX homeobox 1 |
2426 |
0.3 |
| chr3_141464293_141464444 | 0.24 |
Unc5c |
unc-5 netrin receptor C |
848 |
0.47 |
| chr6_148441933_148442246 | 0.24 |
Tmtc1 |
transmembrane and tetratricopeptide repeat containing 1 |
454 |
0.84 |
| chrX_103819527_103819771 | 0.24 |
Slc16a2 |
solute carrier family 16 (monocarboxylic acid transporters), member 2 |
2334 |
0.25 |
| chr1_146492868_146493351 | 0.24 |
Brinp3 |
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
1651 |
0.31 |
| chr5_112153542_112154115 | 0.24 |
1700016B01Rik |
RIKEN cDNA 1700016B01 gene |
2409 |
0.28 |
| chr3_56181373_56181654 | 0.24 |
Nbea |
neurobeachin |
2188 |
0.32 |
| chr1_45312046_45312607 | 0.23 |
Gm47302 |
predicted gene, 47302 |
653 |
0.46 |
| chr6_4506970_4507670 | 0.23 |
Gm43921 |
predicted gene, 43921 |
1233 |
0.34 |
| chr8_25911399_25912280 | 0.23 |
Kcnu1 |
potassium channel, subfamily U, member 1 |
147 |
0.95 |
| chr8_36147717_36147868 | 0.23 |
Gm38414 |
predicted gene, 38414 |
8218 |
0.16 |
| chr11_16753788_16754062 | 0.23 |
Egfr |
epidermal growth factor receptor |
1695 |
0.38 |
| chr13_64040042_64040324 | 0.23 |
Hsd17b3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
22950 |
0.17 |
| chr3_30546274_30547260 | 0.23 |
Mecom |
MDS1 and EVI1 complex locus |
1241 |
0.35 |
| chr15_87546913_87547689 | 0.23 |
Tafa5 |
TAFA chemokine like family member 5 |
3002 |
0.41 |
| chr6_58905319_58905541 | 0.23 |
Nap1l5 |
nucleosome assembly protein 1-like 5 |
1243 |
0.38 |
| chr6_105676979_105678296 | 0.23 |
Cntn4 |
contactin 4 |
23 |
0.52 |
| chr14_97931267_97931418 | 0.23 |
Gm25831 |
predicted gene, 25831 |
36649 |
0.23 |
| chr10_24235368_24236239 | 0.23 |
Moxd1 |
monooxygenase, DBH-like 1 |
12258 |
0.2 |
| chr6_4509880_4510830 | 0.22 |
Gm43921 |
predicted gene, 43921 |
4268 |
0.17 |
| chr9_117318478_117318629 | 0.22 |
Gm17396 |
predicted gene, 17396 |
66017 |
0.12 |
| chr15_16564836_16564987 | 0.22 |
Gm2611 |
predicted gene 2611 |
35651 |
0.19 |
| chr1_190977127_190977297 | 0.22 |
Vash2 |
vasohibin 2 |
1790 |
0.22 |
| chr9_113387561_113387731 | 0.22 |
Gm36485 |
predicted gene, 36485 |
32414 |
0.15 |
| chr15_36960162_36960613 | 0.22 |
Gm34590 |
predicted gene, 34590 |
21523 |
0.14 |
| chr17_75177945_75179637 | 0.22 |
Ltbp1 |
latent transforming growth factor beta binding protein 1 |
19 |
0.98 |
| chr7_113950384_113950535 | 0.22 |
Gm45615 |
predicted gene 45615 |
136439 |
0.04 |
| chr6_44179699_44179850 | 0.22 |
Gm35216 |
predicted gene, 35216 |
96419 |
0.09 |
| chr13_112998291_112999283 | 0.22 |
Mcidas |
multiciliate differentiation and DNA synthesis associated cell cycle protein |
4942 |
0.11 |
| chr10_23345710_23346907 | 0.22 |
Eya4 |
EYA transcriptional coactivator and phosphatase 4 |
3579 |
0.36 |
| chr3_146851868_146852360 | 0.22 |
Ttll7 |
tubulin tyrosine ligase-like family, member 7 |
253 |
0.92 |
| chr19_36734732_36735279 | 0.22 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1648 |
0.41 |
| chr19_34101834_34101985 | 0.22 |
Lipm |
lipase, family member M |
969 |
0.49 |
| chr8_61316532_61316683 | 0.21 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
27735 |
0.16 |
| chr14_57056590_57056818 | 0.21 |
Gja3 |
gap junction protein, alpha 3 |
1019 |
0.5 |
| chr18_82006563_82006720 | 0.21 |
Gm50293 |
predicted gene, 50293 |
10807 |
0.17 |
| chr15_93592923_93593281 | 0.21 |
Prickle1 |
prickle planar cell polarity protein 1 |
2789 |
0.35 |
| chr2_110949491_110950578 | 0.21 |
Ano3 |
anoctamin 3 |
338 |
0.92 |
| chr7_139978745_139979435 | 0.21 |
Spef1l |
sperm flagellar 1 like |
335 |
0.7 |
| chr9_117870768_117871285 | 0.21 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
1558 |
0.39 |
| chr2_113755577_113755728 | 0.21 |
Grem1 |
gremlin 1, DAN family BMP antagonist |
2994 |
0.27 |
| chr3_131566361_131566512 | 0.21 |
Papss1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
1501 |
0.48 |
| chr1_45520505_45520757 | 0.21 |
Gm7063 |
predicted gene 7063 |
123 |
0.96 |
| chr6_141252051_141252954 | 0.21 |
Gm28523 |
predicted gene 28523 |
2493 |
0.28 |
| chr2_78210324_78210475 | 0.21 |
Gm14461 |
predicted gene 14461 |
27148 |
0.25 |
| chr17_86350533_86350917 | 0.21 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
63547 |
0.13 |
| chr13_31810334_31810485 | 0.21 |
Foxc1 |
forkhead box C1 |
3776 |
0.2 |
| chr14_48207086_48207793 | 0.21 |
Peli2 |
pellino 2 |
8723 |
0.15 |
| chr3_151833797_151834654 | 0.21 |
Ptgfr |
prostaglandin F receptor |
70 |
0.97 |
| chr12_95699798_95699986 | 0.21 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
4535 |
0.2 |
| chrX_95166552_95167109 | 0.21 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
291 |
0.93 |
| chr19_56822342_56822651 | 0.21 |
Ccdc186 |
coiled-coil domain containing 186 |
306 |
0.88 |
| chr10_125967796_125968797 | 0.21 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
2043 |
0.45 |
| chr3_151002106_151002257 | 0.21 |
Mir3963 |
microRNA 3963 |
21681 |
0.23 |
| chr14_48654042_48654193 | 0.21 |
Gm23469 |
predicted gene, 23469 |
1618 |
0.2 |
| chr9_62857494_62859103 | 0.21 |
Calml4 |
calmodulin-like 4 |
164 |
0.92 |
| chr12_117245902_117246283 | 0.20 |
Mir153 |
microRNA 153 |
4725 |
0.31 |
| chr17_80772743_80773228 | 0.20 |
Map4k3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
44500 |
0.15 |
| chr16_21207731_21208515 | 0.20 |
Ephb3 |
Eph receptor B3 |
2401 |
0.25 |
| chr15_81189593_81189780 | 0.20 |
Mrtfa |
myocardin related transcription factor A |
836 |
0.42 |
| chr16_70562219_70562370 | 0.20 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
1300 |
0.56 |
| chr7_37342624_37344008 | 0.20 |
6720469O03Rik |
RIKEN cDNA 6720469O03 gene |
23314 |
0.21 |
| chr9_101006061_101006212 | 0.20 |
Pccb |
propionyl Coenzyme A carboxylase, beta polypeptide |
20527 |
0.15 |
| chr15_54479454_54479605 | 0.20 |
Gm20070 |
predicted gene, 20070 |
8371 |
0.21 |
| chr13_63571577_63571772 | 0.20 |
Ptch1 |
patched 1 |
1786 |
0.25 |
| chr10_81903421_81903590 | 0.20 |
Zfp781 |
zinc finger protein 781 |
7394 |
0.1 |
| chr10_6790825_6791573 | 0.20 |
Oprm1 |
opioid receptor, mu 1 |
2100 |
0.4 |
| chr19_25060307_25061291 | 0.20 |
Dock8 |
dedicator of cytokinesis 8 |
8186 |
0.24 |
| chr13_49609766_49610088 | 0.20 |
Ogn |
osteoglycin |
1881 |
0.26 |
| chr2_102551752_102552240 | 0.20 |
Pamr1 |
peptidase domain containing associated with muscle regeneration 1 |
1847 |
0.45 |
| chr13_3889296_3889648 | 0.20 |
Net1 |
neuroepithelial cell transforming gene 1 |
1647 |
0.26 |
| chr15_6390004_6390155 | 0.20 |
Dab2 |
disabled 2, mitogen-responsive phosphoprotein |
41 |
0.98 |
| chr6_136080160_136080311 | 0.20 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
91654 |
0.08 |
| chr1_161879355_161879506 | 0.20 |
Suco |
SUN domain containing ossification factor |
2748 |
0.22 |
| chr10_103145360_103145802 | 0.20 |
Lrriq1 |
leucine-rich repeats and IQ motif containing 1 |
794 |
0.68 |
| chr17_33333481_33333632 | 0.20 |
Zfp81 |
zinc finger protein 81 |
25312 |
0.11 |
| chr17_49309279_49309671 | 0.20 |
Gm17830 |
predicted gene, 17830 |
2766 |
0.26 |
| chr5_106292155_106292306 | 0.20 |
Gm38433 |
predicted gene, 38433 |
20020 |
0.15 |
| chr18_40259056_40260097 | 0.20 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
773 |
0.5 |
| chr18_37652785_37653031 | 0.20 |
Gm8242 |
predicted gene 8242 |
1 |
0.91 |
| chr13_118713631_118713983 | 0.20 |
Fgf10 |
fibroblast growth factor 10 |
892 |
0.52 |
| chr3_131738069_131738640 | 0.20 |
Gm29865 |
predicted gene, 29865 |
21451 |
0.25 |
| chr13_85926958_85927576 | 0.19 |
Gm46396 |
predicted gene, 46396 |
35525 |
0.19 |
| chr7_89405888_89406433 | 0.19 |
Fzd4 |
frizzled class receptor 4 |
1805 |
0.24 |
| chr5_106004428_106004686 | 0.19 |
Gm19566 |
predicted gene, 19566 |
23375 |
0.18 |
| chr7_69490962_69491246 | 0.19 |
Gm38584 |
predicted gene, 38584 |
303 |
0.89 |
| chr15_18819772_18819999 | 0.19 |
Cdh10 |
cadherin 10 |
444 |
0.76 |
| chr12_107927728_107928847 | 0.19 |
Bcl11b |
B cell leukemia/lymphoma 11B |
75127 |
0.11 |
| chr13_8205020_8205313 | 0.19 |
Adarb2 |
adenosine deaminase, RNA-specific, B2 |
2244 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.4 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.1 | 0.2 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.2 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.1 | 0.2 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.0 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.0 | 0.0 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.0 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0002692 | negative regulation of cellular extravasation(GO:0002692) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.0 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:1903276 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0050942 | positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.5 | GO:0052715 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |