Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
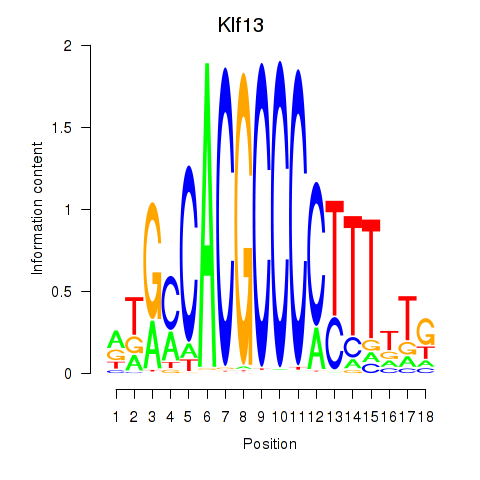
Results for Klf13
Z-value: 0.73
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSMUSG00000052040.9 | Kruppel-like factor 13 |
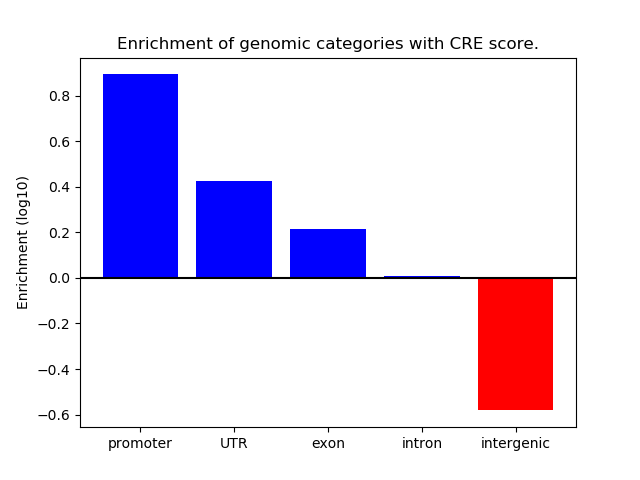
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_63931803_63931954 | Klf13 | 4423 | 0.152193 | -0.30 | 1.9e-02 | Click! |
| chr7_63937849_63938852 | Klf13 | 565 | 0.672893 | -0.30 | 1.9e-02 | Click! |
| chr7_63934435_63935180 | Klf13 | 1494 | 0.304406 | -0.30 | 2.1e-02 | Click! |
| chr7_63941200_63941688 | Klf13 | 2529 | 0.196957 | 0.25 | 5.6e-02 | Click! |
| chr7_63941730_63941881 | Klf13 | 2890 | 0.181158 | 0.21 | 1.1e-01 | Click! |
Activity of the Klf13 motif across conditions
Conditions sorted by the z-value of the Klf13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
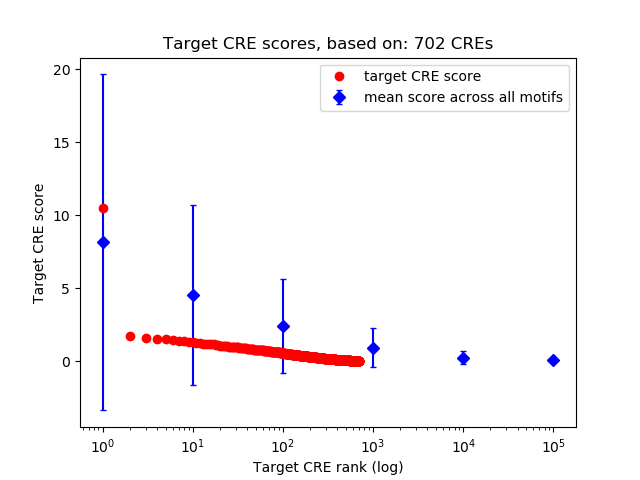
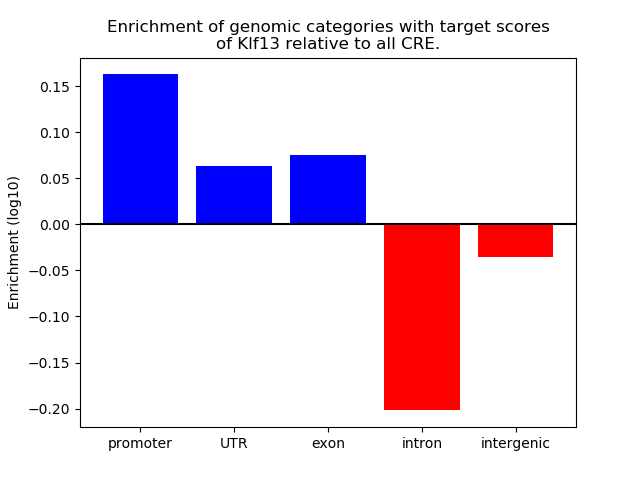
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_139020697_139021233 | 10.52 |
Gm45613 |
predicted gene 45613 |
34 |
0.98 |
| chr8_71558187_71559532 | 1.72 |
Tmem221 |
transmembrane protein 221 |
12 |
0.94 |
| chr13_60737782_60738368 | 1.56 |
Dapk1 |
death associated protein kinase 1 |
15921 |
0.15 |
| chr3_41522196_41522994 | 1.53 |
2400006E01Rik |
RIKEN cDNA 2400006E01 gene |
622 |
0.65 |
| chr15_80598900_80599687 | 1.50 |
Grap2 |
GRB2-related adaptor protein 2 |
24190 |
0.13 |
| chr14_63164156_63165244 | 1.43 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
225 |
0.91 |
| chr17_56663591_56664733 | 1.40 |
Ranbp3 |
RAN binding protein 3 |
9132 |
0.11 |
| chr17_56424927_56425952 | 1.38 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
22 |
0.97 |
| chr6_119416453_119417745 | 1.35 |
Adipor2 |
adiponectin receptor 2 |
376 |
0.88 |
| chrX_137118132_137120673 | 1.30 |
Esx1 |
extraembryonic, spermatogenesis, homeobox 1 |
769 |
0.36 |
| chr10_80319682_80319974 | 1.28 |
2310011J03Rik |
RIKEN cDNA 2310011J03 gene |
709 |
0.38 |
| chr8_104042374_104042662 | 1.23 |
Gm8748 |
predicted gene 8748 |
28631 |
0.16 |
| chr5_74703847_74705020 | 1.19 |
Gm15984 |
predicted gene 15984 |
1413 |
0.31 |
| chr2_72242133_72242510 | 1.18 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
27 |
0.97 |
| chr12_117447118_117447786 | 1.18 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
69027 |
0.12 |
| chr2_163356404_163356958 | 1.17 |
Tox2 |
TOX high mobility group box family member 2 |
36303 |
0.11 |
| chr6_51193157_51193734 | 1.16 |
Mir148a |
microRNA 148a |
76465 |
0.1 |
| chr7_19180970_19181875 | 1.15 |
Mir330 |
microRNA 330 |
43 |
0.54 |
| chr9_65128098_65129057 | 1.13 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
12612 |
0.13 |
| chr17_29841641_29842774 | 1.06 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
214 |
0.93 |
| chr4_141041263_141041822 | 1.05 |
Crocc |
ciliary rootlet coiled-coil, rootletin |
12119 |
0.11 |
| chr5_72224662_72225245 | 1.05 |
Atp10d |
ATPase, class V, type 10D |
21600 |
0.17 |
| chr7_45515256_45516241 | 1.03 |
Tulp2 |
tubby-like protein 2 |
60 |
0.91 |
| chr2_135536052_135537171 | 1.02 |
9630028H03Rik |
RIKEN cDNA 9630028H03 gene |
46685 |
0.17 |
| chr2_28369440_28369591 | 1.00 |
Gm13373 |
predicted gene 13373 |
45465 |
0.12 |
| chr7_135863729_135863975 | 1.00 |
Gm45241 |
predicted gene 45241 |
9404 |
0.16 |
| chr15_75708830_75709401 | 1.00 |
Rhpn1 |
rhophilin, Rho GTPase binding protein 1 |
1624 |
0.26 |
| chr15_101221041_101222412 | 0.98 |
AU021063 |
expressed sequence AU021063 |
479 |
0.63 |
| chr15_76242501_76243437 | 0.98 |
Parp10 |
poly (ADP-ribose) polymerase family, member 10 |
451 |
0.59 |
| chr19_56474746_56475273 | 0.97 |
Plekhs1 |
pleckstrin homology domain containing, family S member 1 |
3892 |
0.23 |
| chr10_44038032_44038602 | 0.96 |
Crybg1 |
crystallin beta-gamma domain containing 1 |
20461 |
0.18 |
| chr7_45800366_45800681 | 0.95 |
Cyth2 |
cytohesin 2 |
10365 |
0.07 |
| chr10_61491946_61492200 | 0.93 |
Gm47595 |
predicted gene, 47595 |
4217 |
0.12 |
| chr16_44015370_44016774 | 0.92 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr1_195091807_195092906 | 0.92 |
Cd46 |
CD46 antigen, complement regulatory protein |
107 |
0.94 |
| chr11_69964787_69965749 | 0.89 |
Cldn7 |
claudin 7 |
128 |
0.87 |
| chr2_29546441_29547087 | 0.89 |
Med27 |
mediator complex subunit 27 |
41893 |
0.12 |
| chr8_70477508_70477693 | 0.88 |
Klhl26 |
kelch-like 26 |
632 |
0.51 |
| chr7_27449664_27449998 | 0.88 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
1770 |
0.18 |
| chr3_96102132_96102636 | 0.87 |
Otud7b |
OTU domain containing 7B |
2143 |
0.17 |
| chr13_98689529_98690460 | 0.85 |
Tmem171 |
transmembrane protein 171 |
4774 |
0.15 |
| chr8_120508825_120510253 | 0.84 |
Gm26971 |
predicted gene, 26971 |
11076 |
0.13 |
| chr14_76585926_76586134 | 0.84 |
Serp2 |
stress-associated endoplasmic reticulum protein family member 2 |
29141 |
0.16 |
| chr4_124899678_124899829 | 0.82 |
Epha10 |
Eph receptor A10 |
351 |
0.78 |
| chr7_15954786_15955942 | 0.82 |
Nop53 |
NOP53 ribosome biogenesis factor |
9290 |
0.1 |
| chr10_93833520_93834579 | 0.81 |
Usp44 |
ubiquitin specific peptidase 44 |
231 |
0.91 |
| chr5_118482858_118484042 | 0.81 |
Gm15754 |
predicted gene 15754 |
3517 |
0.25 |
| chr16_90739931_90740383 | 0.80 |
Mrap |
melanocortin 2 receptor accessory protein |
1833 |
0.28 |
| chr7_45340898_45341049 | 0.80 |
Ppfia3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
187 |
0.83 |
| chr11_117425952_117426103 | 0.80 |
Gm11732 |
predicted gene 11732 |
151 |
0.94 |
| chr5_143924982_143925490 | 0.79 |
Gm42421 |
predicted gene, 42421 |
2939 |
0.15 |
| chr4_152107258_152108639 | 0.79 |
Plekhg5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
219 |
0.89 |
| chr12_24707675_24708235 | 0.79 |
Rrm2 |
ribonucleotide reductase M2 |
286 |
0.87 |
| chr7_133114831_133116529 | 0.77 |
Ctbp2 |
C-terminal binding protein 2 |
2981 |
0.2 |
| chr1_87270545_87271387 | 0.77 |
Efhd1os |
EF hand domain containing 1, opposite strand |
5792 |
0.12 |
| chr8_94182622_94183855 | 0.77 |
Gm39228 |
predicted gene, 39228 |
51 |
0.95 |
| chr1_136008028_136008415 | 0.76 |
Tmem9 |
transmembrane protein 9 |
71 |
0.95 |
| chr9_109876011_109877009 | 0.75 |
Cdc25a |
cell division cycle 25A |
204 |
0.9 |
| chr7_80156798_80157153 | 0.74 |
Gm45202 |
predicted gene 45202 |
1806 |
0.23 |
| chr11_117175159_117175396 | 0.74 |
Sec14l1 |
SEC14-like lipid binding 1 |
20208 |
0.13 |
| chr14_76149853_76150359 | 0.74 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
39215 |
0.15 |
| chr2_32750921_32752008 | 0.73 |
Tor2a |
torsin family 2, member A |
5770 |
0.08 |
| chr14_65149139_65150455 | 0.73 |
Extl3 |
exostosin-like glycosyltransferase 3 |
58 |
0.97 |
| chr9_20990858_20991262 | 0.73 |
Mrpl4 |
mitochondrial ribosomal protein L4 |
11678 |
0.07 |
| chr10_120061270_120061896 | 0.72 |
Helb |
helicase (DNA) B |
30626 |
0.18 |
| chr19_43584864_43585184 | 0.72 |
Gm34636 |
predicted gene, 34636 |
11667 |
0.11 |
| chr9_107288899_107290229 | 0.70 |
Mapkapk3 |
mitogen-activated protein kinase-activated protein kinase 3 |
183 |
0.89 |
| chr17_13668170_13669126 | 0.70 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
243 |
0.9 |
| chr13_58007872_58008023 | 0.69 |
Mir874 |
microRNA 874 |
15253 |
0.21 |
| chr4_155468952_155469786 | 0.69 |
Tmem52 |
transmembrane protein 52 |
222 |
0.87 |
| chr1_165763123_165763707 | 0.68 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
331 |
0.77 |
| chr17_47802972_47803123 | 0.68 |
Tfeb |
transcription factor EB |
13606 |
0.12 |
| chr2_93461586_93463194 | 0.68 |
Cd82 |
CD82 antigen |
4 |
0.98 |
| chr15_83496013_83496555 | 0.67 |
Ttll1 |
tubulin tyrosine ligase-like 1 |
14509 |
0.12 |
| chr1_182585252_182585888 | 0.67 |
Capn8 |
calpain 8 |
20532 |
0.16 |
| chr14_75069008_75069423 | 0.67 |
Rubcnl |
RUN and cysteine rich domain containing beclin 1 interacting protein like |
32593 |
0.14 |
| chr3_39219846_39220607 | 0.65 |
Gm43008 |
predicted gene 43008 |
2380 |
0.41 |
| chr1_134747452_134748117 | 0.65 |
Syt2 |
synaptotagmin II |
793 |
0.59 |
| chr10_43666883_43667818 | 0.65 |
Gm34481 |
predicted gene, 34481 |
8032 |
0.13 |
| chr2_163823554_163824237 | 0.65 |
Ccn5 |
cellular communication network factor 5 |
3006 |
0.24 |
| chr10_81168389_81168622 | 0.65 |
Pias4 |
protein inhibitor of activated STAT 4 |
582 |
0.47 |
| chr17_47909108_47910390 | 0.64 |
Gm15556 |
predicted gene 15556 |
12629 |
0.13 |
| chr1_160746842_160746993 | 0.63 |
Gm38126 |
predicted gene, 38126 |
8770 |
0.12 |
| chr6_142788560_142788823 | 0.63 |
Sult6b2 |
sulfotransferase family 6B, member 2 |
15775 |
0.16 |
| chr15_99113363_99113593 | 0.63 |
Spats2 |
spermatogenesis associated, serine-rich 2 |
12438 |
0.09 |
| chr8_94781701_94782201 | 0.63 |
Cx3cl1 |
chemokine (C-X3-C motif) ligand 1 |
9726 |
0.11 |
| chr13_112439365_112440345 | 0.62 |
Gm48879 |
predicted gene, 48879 |
3259 |
0.19 |
| chr16_90400287_90400987 | 0.62 |
Hunk |
hormonally upregulated Neu-associated kinase |
93 |
0.97 |
| chr7_49496645_49497633 | 0.62 |
Gm38059 |
predicted gene, 38059 |
27427 |
0.2 |
| chr11_59786838_59787832 | 0.61 |
Pld6 |
phospholipase D family, member 6 |
307 |
0.8 |
| chr9_114527981_114528394 | 0.61 |
C130032M10Rik |
RIKEN cDNA C130032M10 gene |
12490 |
0.16 |
| chr8_117335167_117335696 | 0.61 |
Cmip |
c-Maf inducing protein |
13739 |
0.25 |
| chr5_122059493_122060141 | 0.60 |
Cux2 |
cut-like homeobox 2 |
9715 |
0.14 |
| chr2_167343559_167343841 | 0.59 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
5483 |
0.21 |
| chr4_138469035_138469593 | 0.58 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
15000 |
0.15 |
| chr4_132796191_132796588 | 0.58 |
Themis2 |
thymocyte selection associated family member 2 |
2 |
0.96 |
| chr2_168158942_168159659 | 0.58 |
E130018N17Rik |
RIKEN cDNA E130018N17 gene |
4787 |
0.13 |
| chr11_119237747_119238894 | 0.57 |
Ccdc40 |
coiled-coil domain containing 40 |
398 |
0.77 |
| chr11_117475760_117476250 | 0.56 |
Gm34418 |
predicted gene, 34418 |
6592 |
0.15 |
| chr11_78435586_78436201 | 0.55 |
Gm11195 |
predicted gene 11195 |
8214 |
0.09 |
| chr19_29325359_29325923 | 0.55 |
Insl6 |
insulin-like 6 |
285 |
0.88 |
| chr9_120581966_120582700 | 0.55 |
Gm47062 |
predicted gene, 47062 |
2198 |
0.19 |
| chr13_23317523_23318359 | 0.54 |
4930586N03Rik |
RIKEN cDNA 4930586N03 gene |
4416 |
0.1 |
| chr14_54958124_54958655 | 0.54 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
482 |
0.52 |
| chr9_107526345_107527662 | 0.53 |
Cacna2d2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
346 |
0.68 |
| chr11_34031937_34032124 | 0.53 |
Lcp2 |
lymphocyte cytosolic protein 2 |
14890 |
0.2 |
| chr19_10421963_10422290 | 0.53 |
Gm50268 |
predicted gene, 50268 |
17771 |
0.13 |
| chr15_84450356_84451064 | 0.52 |
Shisal1 |
shisa like 1 |
3613 |
0.28 |
| chr13_92593987_92594157 | 0.52 |
Serinc5 |
serine incorporator 5 |
17019 |
0.18 |
| chr7_98158554_98158943 | 0.52 |
Capn5 |
calpain 5 |
3462 |
0.18 |
| chr17_47737108_47738630 | 0.52 |
Tfeb |
transcription factor EB |
59 |
0.95 |
| chr14_20452251_20452547 | 0.52 |
Cfap70 |
cilia and flagella associated protein 70 |
173 |
0.92 |
| chr9_44515217_44515909 | 0.52 |
Cxcr5 |
chemokine (C-X-C motif) receptor 5 |
10858 |
0.07 |
| chr2_30985262_30985875 | 0.52 |
Usp20 |
ubiquitin specific peptidase 20 |
3139 |
0.16 |
| chr5_101665386_101665820 | 0.51 |
Nkx6-1 |
NK6 homeobox 1 |
607 |
0.74 |
| chr12_108004788_108005664 | 0.51 |
Bcl11b |
B cell leukemia/lymphoma 11B |
1624 |
0.51 |
| chr13_13358078_13358229 | 0.51 |
Gpr137b |
G protein-coupled receptor 137B |
9652 |
0.11 |
| chrX_20659340_20659491 | 0.51 |
Uba1 |
ubiquitin-like modifier activating enzyme 1 |
12 |
0.97 |
| chr3_33955028_33955284 | 0.51 |
4930502C17Rik |
RIKEN cDNA 4930502C17 gene |
1044 |
0.44 |
| chr2_73589836_73590674 | 0.50 |
Chrna1os |
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle), opposite strand |
5395 |
0.15 |
| chr10_80569426_80569836 | 0.50 |
Klf16 |
Kruppel-like factor 16 |
7690 |
0.08 |
| chr6_90989200_90989476 | 0.50 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
653 |
0.72 |
| chr7_99827539_99828752 | 0.50 |
Neu3 |
neuraminidase 3 |
272 |
0.86 |
| chr7_45797605_45798943 | 0.49 |
Lmtk3 |
lemur tyrosine kinase 3 |
10526 |
0.07 |
| chr4_43493224_43493378 | 0.49 |
Ccdc107 |
coiled-coil domain containing 107 |
61 |
0.71 |
| chr10_42715047_42716006 | 0.46 |
Gm15200 |
predicted gene 15200 |
17008 |
0.16 |
| chr5_144357138_144358729 | 0.45 |
Dmrt1i |
Dmrt1 interacting ncRNA |
10 |
0.7 |
| chr16_10357804_10358541 | 0.45 |
Gm1600 |
predicted gene 1600 |
10581 |
0.15 |
| chr15_36647405_36647556 | 0.45 |
Gm6704 |
predicted gene 6704 |
17611 |
0.12 |
| chr2_57102068_57103416 | 0.45 |
Nr4a2 |
nuclear receptor subfamily 4, group A, member 2 |
10330 |
0.2 |
| chr15_11741805_11742688 | 0.45 |
Gm41271 |
predicted gene, 41271 |
38543 |
0.18 |
| chr17_34093589_34094165 | 0.45 |
BC051537 |
cDNA sequence BC051537 |
1405 |
0.13 |
| chr5_123171494_123172535 | 0.44 |
Hpd |
4-hydroxyphenylpyruvic acid dioxygenase |
897 |
0.35 |
| chr2_180642344_180643352 | 0.44 |
Tcfl5 |
transcription factor-like 5 (basic helix-loop-helix) |
140 |
0.94 |
| chr11_117685805_117686516 | 0.44 |
Tnrc6c |
trinucleotide repeat containing 6C |
31347 |
0.13 |
| chr14_58383123_58383274 | 0.44 |
Gm33321 |
predicted gene, 33321 |
82216 |
0.1 |
| chr13_37508920_37510039 | 0.43 |
Gm29590 |
predicted gene 29590 |
239 |
0.85 |
| chr7_45437080_45437256 | 0.43 |
Ruvbl2 |
RuvB-like protein 2 |
928 |
0.24 |
| chr8_120693953_120694425 | 0.42 |
Gm33142 |
predicted gene, 33142 |
8790 |
0.11 |
| chr17_56818209_56818531 | 0.42 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
12569 |
0.12 |
| chr1_135778673_135779502 | 0.42 |
Tnni1 |
troponin I, skeletal, slow 1 |
347 |
0.84 |
| chr5_106470004_106471103 | 0.42 |
Gm26872 |
predicted gene, 26872 |
454 |
0.81 |
| chr17_24657836_24658944 | 0.42 |
Npw |
neuropeptide W |
67 |
0.92 |
| chr2_170154822_170155722 | 0.41 |
Zfp217 |
zinc finger protein 217 |
7169 |
0.29 |
| chr2_32083168_32084169 | 0.41 |
Fam78a |
family with sequence similarity 78, member A |
115 |
0.94 |
| chr15_100636108_100637173 | 0.41 |
Smagp |
small cell adhesion glycoprotein |
51 |
0.94 |
| chr10_81380285_81380531 | 0.41 |
Fzr1 |
fizzy and cell division cycle 20 related 1 |
1892 |
0.11 |
| chr7_25619112_25619835 | 0.41 |
Dmac2 |
distal membrane arm assembly complex 2 |
20 |
0.94 |
| chr7_18962222_18963745 | 0.40 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
24417 |
0.07 |
| chr4_32834117_32834411 | 0.40 |
Ankrd6 |
ankyrin repeat domain 6 |
11463 |
0.18 |
| chr7_29118392_29118870 | 0.40 |
Ryr1 |
ryanodine receptor 1, skeletal muscle |
6384 |
0.09 |
| chr15_82293133_82293794 | 0.40 |
Wbp2nl |
WBP2 N-terminal like |
5491 |
0.09 |
| chr16_30512552_30512703 | 0.39 |
Tmem44 |
transmembrane protein 44 |
28282 |
0.16 |
| chr17_56756624_56758021 | 0.39 |
Nrtn |
neurturin |
208 |
0.88 |
| chr5_41763315_41764785 | 0.39 |
Nkx3-2 |
NK3 homeobox 2 |
451 |
0.86 |
| chr11_69326020_69327207 | 0.39 |
Kcnab3 |
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
355 |
0.68 |
| chr7_145302223_145303241 | 0.39 |
Mrgprf |
MAS-related GPR, member F |
1833 |
0.35 |
| chr2_119334790_119335432 | 0.39 |
Dll4 |
delta like canonical Notch ligand 4 |
6573 |
0.13 |
| chr11_74536337_74536639 | 0.39 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
13737 |
0.2 |
| chr17_46620556_46620966 | 0.39 |
Ptk7 |
PTK7 protein tyrosine kinase 7 |
8743 |
0.1 |
| chr9_64022235_64023090 | 0.39 |
Smad6 |
SMAD family member 6 |
603 |
0.67 |
| chr10_45291931_45292275 | 0.38 |
Gm15934 |
predicted gene 15934 |
2679 |
0.18 |
| chr7_16313330_16314024 | 0.38 |
Bbc3 |
BCL2 binding component 3 |
160 |
0.92 |
| chr8_71665120_71666309 | 0.38 |
Unc13a |
unc-13 homolog A |
2368 |
0.14 |
| chr7_44350602_44354420 | 0.38 |
Shank1 |
SH3 and multiple ankyrin repeat domains 1 |
1749 |
0.15 |
| chr10_67126563_67127770 | 0.38 |
Jmjd1c |
jumonji domain containing 1C |
92 |
0.97 |
| chr13_95524182_95525409 | 0.38 |
F2rl1 |
coagulation factor II (thrombin) receptor-like 1 |
432 |
0.8 |
| chr11_99131599_99132803 | 0.38 |
Ccr7 |
chemokine (C-C motif) receptor 7 |
22876 |
0.13 |
| chr16_17619128_17619354 | 0.37 |
Smpd4 |
sphingomyelin phosphodiesterase 4 |
113 |
0.94 |
| chr7_30183828_30184670 | 0.37 |
Cox7a1 |
cytochrome c oxidase subunit 7A1 |
55 |
0.92 |
| chr4_140770313_140770939 | 0.37 |
Padi4 |
peptidyl arginine deiminase, type IV |
3610 |
0.16 |
| chr6_118593001_118594277 | 0.37 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1787 |
0.35 |
| chr7_49350531_49350896 | 0.36 |
Gm44913 |
predicted gene 44913 |
4619 |
0.25 |
| chr17_49900579_49901205 | 0.36 |
Gm16554 |
predicted gene 16554 |
4925 |
0.2 |
| chr5_100353191_100353824 | 0.36 |
Gm8091 |
predicted gene 8091 |
1612 |
0.36 |
| chr8_70630745_70631808 | 0.36 |
Gdf15 |
growth differentiation factor 15 |
819 |
0.39 |
| chr11_47377104_47378325 | 0.36 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
1808 |
0.52 |
| chr13_62857070_62857298 | 0.36 |
Fbp2 |
fructose bisphosphatase 2 |
875 |
0.48 |
| chr2_158153509_158154452 | 0.36 |
Tgm2 |
transglutaminase 2, C polypeptide |
7544 |
0.16 |
| chr18_82906600_82906751 | 0.35 |
Zfp516 |
zinc finger protein 516 |
3988 |
0.17 |
| chr8_70329770_70331438 | 0.35 |
Gdf1 |
growth differentiation factor 1 |
787 |
0.45 |
| chr11_117836263_117836628 | 0.35 |
Afmid |
arylformamidase |
4172 |
0.1 |
| chr10_119614719_119615429 | 0.35 |
Gm47027 |
predicted gene, 47027 |
2646 |
0.29 |
| chr10_61906333_61906511 | 0.35 |
Col13a1 |
collagen, type XIII, alpha 1 |
28030 |
0.17 |
| chr7_29318511_29319550 | 0.34 |
Gm44632 |
predicted gene 44632 |
372 |
0.77 |
| chr11_120350589_120350893 | 0.34 |
0610009L18Rik |
RIKEN cDNA 0610009L18 gene |
2063 |
0.13 |
| chr12_111943340_111944555 | 0.34 |
5033406O09Rik |
RIKEN cDNA 5033406O09 gene |
199 |
0.89 |
| chr6_120187572_120188939 | 0.34 |
Ninj2 |
ninjurin 2 |
5568 |
0.22 |
| chr10_39501612_39501987 | 0.33 |
Fyn |
Fyn proto-oncogene |
9940 |
0.21 |
| chr1_133168432_133168872 | 0.33 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
4442 |
0.19 |
| chr18_84070282_84070920 | 0.33 |
Tshz1 |
teashirt zinc finger family member 1 |
14474 |
0.16 |
| chr3_108558558_108558709 | 0.32 |
Gm43435 |
predicted gene 43435 |
3166 |
0.1 |
| chr3_149076913_149077569 | 0.32 |
Gm25127 |
predicted gene, 25127 |
48309 |
0.14 |
| chr12_105705647_105706505 | 0.32 |
Ak7 |
adenylate kinase 7 |
100 |
0.96 |
| chr17_45526364_45526864 | 0.32 |
Tcte1 |
t-complex-associated testis expressed 1 |
3180 |
0.13 |
| chr13_98968150_98968301 | 0.32 |
Gm47052 |
predicted gene, 47052 |
5153 |
0.13 |
| chr2_167538066_167539536 | 0.32 |
Snai1 |
snail family zinc finger 1 |
606 |
0.61 |
| chr11_35838085_35838789 | 0.32 |
Rars |
arginyl-tRNA synthetase |
3931 |
0.2 |
| chr18_63859767_63860372 | 0.31 |
Gm19076 |
predicted gene, 19076 |
38962 |
0.15 |
| chr11_108222674_108223259 | 0.31 |
Gm11655 |
predicted gene 11655 |
41116 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.3 | 1.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.7 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.2 | 1.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 1.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 0.8 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.4 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.8 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.8 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.5 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.1 | 0.2 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.2 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.3 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 1.1 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.7 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.2 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.2 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.0 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.0 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 0.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.5 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 1.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |