Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
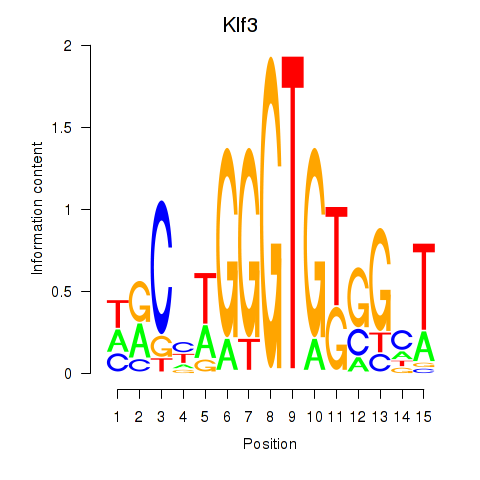
Results for Klf3
Z-value: 0.58
Transcription factors associated with Klf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf3
|
ENSMUSG00000029178.8 | Kruppel-like factor 3 (basic) |
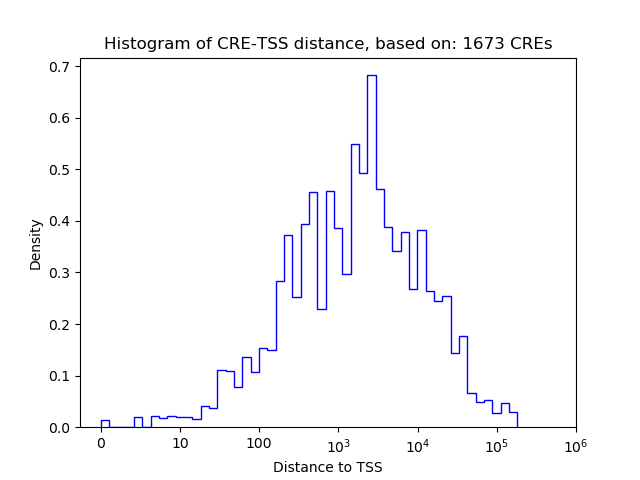
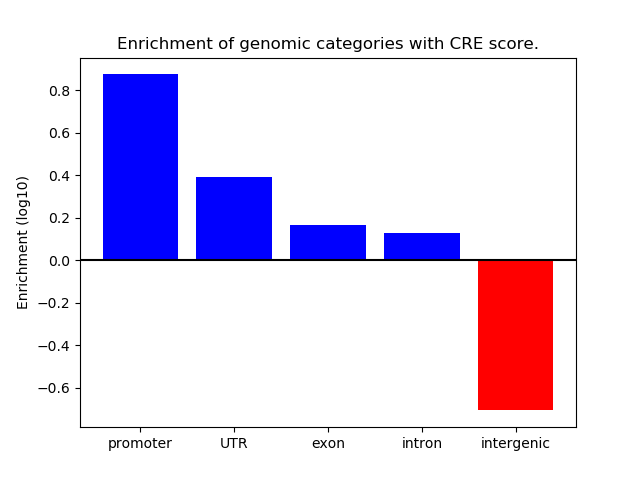
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_64809367_64809770 | Klf3 | 2771 | 0.207271 | 0.29 | 2.4e-02 | Click! |
| chr5_64802596_64803261 | Klf3 | 460 | 0.576944 | -0.26 | 4.9e-02 | Click! |
| chr5_64809804_64809955 | Klf3 | 2460 | 0.224155 | 0.25 | 5.1e-02 | Click! |
| chr5_64813848_64814093 | Klf3 | 1586 | 0.322668 | 0.24 | 7.0e-02 | Click! |
| chr5_64781139_64781290 | Klf3 | 22174 | 0.133905 | 0.14 | 2.7e-01 | Click! |
Activity of the Klf3 motif across conditions
Conditions sorted by the z-value of the Klf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
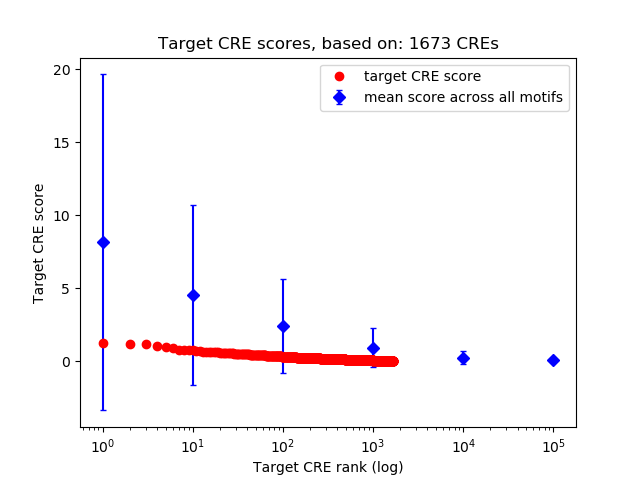
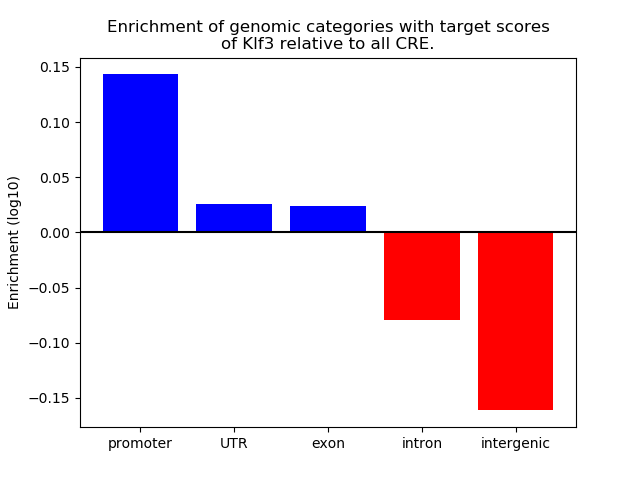
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_102360845_102363484 | 1.22 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
1540 |
0.24 |
| chr4_155893812_155895517 | 1.20 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2352 |
0.11 |
| chr11_90677181_90677343 | 1.16 |
Tom1l1 |
target of myb1-like 1 (chicken) |
10317 |
0.2 |
| chr18_67573133_67573367 | 1.05 |
Gm17669 |
predicted gene, 17669 |
10856 |
0.17 |
| chr16_23058440_23059079 | 0.99 |
Kng1 |
kininogen 1 |
366 |
0.69 |
| chr2_74713120_74713846 | 0.91 |
Hoxd3os1 |
homeobox D3, opposite strand 1 |
957 |
0.21 |
| chr17_56241459_56242409 | 0.77 |
A230051N06Rik |
RIKEN cDNA A230051N06 gene |
218 |
0.81 |
| chr7_45047563_45050465 | 0.76 |
Prr12 |
proline rich 12 |
3867 |
0.07 |
| chr19_47017426_47018238 | 0.75 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
2679 |
0.17 |
| chr17_48450856_48451498 | 0.75 |
Tspo2 |
translocator protein 2 |
315 |
0.83 |
| chr14_66344363_66345813 | 0.72 |
Stmn4 |
stathmin-like 4 |
707 |
0.65 |
| chr10_127167223_127168270 | 0.69 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
1887 |
0.15 |
| chr11_102364387_102365146 | 0.67 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
481 |
0.67 |
| chr6_90551160_90552107 | 0.64 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
785 |
0.56 |
| chr17_48272521_48272834 | 0.63 |
Treml4 |
triggering receptor expressed on myeloid cells-like 4 |
238 |
0.88 |
| chr6_52229552_52230397 | 0.61 |
Mir196b |
microRNA 196b |
191 |
0.57 |
| chr7_79502506_79503035 | 0.61 |
Mir9-3 |
microRNA 9-3 |
2494 |
0.13 |
| chr2_27981777_27982295 | 0.61 |
Col5a1 |
collagen, type V, alpha 1 |
35405 |
0.15 |
| chr1_130732649_130733832 | 0.60 |
AA986860 |
expressed sequence AA986860 |
1130 |
0.29 |
| chr1_172894740_172894971 | 0.59 |
Apcs |
serum amyloid P-component |
186 |
0.93 |
| chr17_88460455_88460676 | 0.57 |
Foxn2 |
forkhead box N2 |
19790 |
0.17 |
| chr1_74949307_74952042 | 0.56 |
Ihh |
Indian hedgehog |
768 |
0.5 |
| chr19_5842574_5845856 | 0.56 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1044 |
0.25 |
| chr11_6413239_6413536 | 0.56 |
Ppia |
peptidylprolyl isomerase A |
2056 |
0.17 |
| chr7_101422402_101423202 | 0.55 |
Pde2a |
phosphodiesterase 2A, cGMP-stimulated |
956 |
0.45 |
| chr13_54607077_54607512 | 0.55 |
Cltb |
clathrin, light polypeptide (Lcb) |
3998 |
0.12 |
| chr5_139382341_139382984 | 0.54 |
Gpr146 |
G protein-coupled receptor 146 |
2081 |
0.2 |
| chr7_27448641_27448983 | 0.53 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
751 |
0.39 |
| chrX_7640827_7641363 | 0.53 |
Syp |
synaptophysin |
1334 |
0.21 |
| chr6_83068298_83071797 | 0.52 |
Tlx2 |
T cell leukemia, homeobox 2 |
178 |
0.81 |
| chr7_126702563_126704731 | 0.51 |
Coro1a |
coronin, actin binding protein 1A |
473 |
0.55 |
| chr16_42275550_42276525 | 0.50 |
Gap43 |
growth associated protein 43 |
35 |
0.98 |
| chr5_123150083_123151176 | 0.50 |
Setd1b |
SET domain containing 1B |
7672 |
0.08 |
| chr17_85686512_85689764 | 0.49 |
Six2 |
sine oculis-related homeobox 2 |
116 |
0.96 |
| chr5_124451141_124451723 | 0.49 |
Kmt5a |
lysine methyltransferase 5A |
261 |
0.84 |
| chr7_19022230_19023942 | 0.49 |
Foxa3 |
forkhead box A3 |
452 |
0.57 |
| chr2_127362687_127364202 | 0.49 |
Adra2b |
adrenergic receptor, alpha 2b |
158 |
0.94 |
| chr13_43261355_43261506 | 0.48 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
41975 |
0.15 |
| chr17_78248797_78249192 | 0.48 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
11222 |
0.18 |
| chr15_100402856_100403333 | 0.47 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
149 |
0.92 |
| chr5_112345255_112345758 | 0.47 |
Hps4 |
HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 |
2406 |
0.14 |
| chr19_7421074_7423945 | 0.47 |
Mir6991 |
microRNA 6991 |
64 |
0.94 |
| chr17_12378732_12379615 | 0.46 |
Plg |
plasminogen |
514 |
0.75 |
| chr15_102976255_102978005 | 0.46 |
Hoxc9 |
homeobox C9 |
98 |
0.92 |
| chr13_21776625_21776982 | 0.44 |
H1f5 |
H1.5 linker histone, cluster member |
3822 |
0.06 |
| chr4_152349775_152350796 | 0.44 |
Gm13645 |
predicted gene 13645 |
3356 |
0.16 |
| chr9_74865733_74868961 | 0.44 |
Onecut1 |
one cut domain, family member 1 |
863 |
0.54 |
| chr3_66486206_66486617 | 0.44 |
Gm17952 |
predicted gene, 17952 |
77140 |
0.1 |
| chr19_6398611_6400187 | 0.44 |
Rasgrp2 |
RAS, guanyl releasing protein 2 |
57 |
0.93 |
| chr2_24951893_24952139 | 0.43 |
Zmynd19 |
zinc finger, MYND domain containing 19 |
1806 |
0.16 |
| chr17_27183697_27183996 | 0.43 |
Lemd2 |
LEM domain containing 2 |
9634 |
0.1 |
| chrX_74426690_74427045 | 0.43 |
Ikbkg |
inhibitor of kappaB kinase gamma |
1035 |
0.31 |
| chr16_91219818_91221532 | 0.42 |
Gm49614 |
predicted gene, 49614 |
2251 |
0.2 |
| chr5_73246539_73246690 | 0.41 |
Fryl |
FRY like transcription coactivator |
9811 |
0.11 |
| chr7_25674373_25675380 | 0.41 |
Tmem91 |
transmembrane protein 91 |
171 |
0.89 |
| chr10_81024569_81025640 | 0.41 |
Gm16099 |
predicted gene 16099 |
21 |
0.8 |
| chr10_81091601_81092278 | 0.41 |
Creb3l3 |
cAMP responsive element binding protein 3-like 3 |
33 |
0.94 |
| chr2_102867441_102867713 | 0.41 |
Gm23731 |
predicted gene, 23731 |
30383 |
0.18 |
| chr7_49398916_49399175 | 0.40 |
Nav2 |
neuron navigator 2 |
34324 |
0.18 |
| chr5_22341020_22341652 | 0.40 |
Reln |
reelin |
3354 |
0.18 |
| chr11_17007546_17008653 | 0.40 |
Plek |
pleckstrin |
603 |
0.7 |
| chr4_125122525_125123430 | 0.40 |
Zc3h12a |
zinc finger CCCH type containing 12A |
457 |
0.77 |
| chr15_75892413_75892732 | 0.40 |
Naprt |
nicotinate phosphoribosyltransferase |
4 |
0.94 |
| chr11_84520959_84524590 | 0.40 |
Lhx1 |
LIM homeobox protein 1 |
63 |
0.97 |
| chr18_64265796_64267074 | 0.39 |
St8sia3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
47 |
0.97 |
| chr8_71700550_71701837 | 0.39 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
596 |
0.52 |
| chr16_49775252_49775761 | 0.39 |
Gm15518 |
predicted gene 15518 |
23364 |
0.2 |
| chr5_139211825_139211976 | 0.39 |
Sun1 |
Sad1 and UNC84 domain containing 1 |
41 |
0.97 |
| chr11_77783639_77784980 | 0.39 |
Gm10277 |
predicted gene 10277 |
3438 |
0.18 |
| chr8_123410787_123412789 | 0.38 |
Tubb3 |
tubulin, beta 3 class III |
198 |
0.84 |
| chr1_166130460_166130958 | 0.37 |
Gpa33 |
glycoprotein A33 (transmembrane) |
242 |
0.91 |
| chr2_121357946_121359451 | 0.37 |
Ckmt1 |
creatine kinase, mitochondrial 1, ubiquitous |
32 |
0.95 |
| chr10_95880189_95880470 | 0.36 |
Gm47724 |
predicted gene, 47724 |
9530 |
0.13 |
| chr1_63655959_63656110 | 0.36 |
Gm12748 |
predicted gene 12748 |
15434 |
0.15 |
| chr1_132384306_132384539 | 0.36 |
Gm15849 |
predicted gene 15849 |
3293 |
0.17 |
| chr7_16275527_16275803 | 0.36 |
Ccdc9 |
coiled-coil domain containing 9 |
106 |
0.93 |
| chr6_108659051_108660392 | 0.36 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
908 |
0.41 |
| chr9_44396453_44399065 | 0.35 |
Slc37a4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
417 |
0.53 |
| chr7_98493200_98493540 | 0.35 |
Lrrc32 |
leucine rich repeat containing 32 |
852 |
0.49 |
| chr3_8509825_8511666 | 0.35 |
Stmn2 |
stathmin-like 2 |
1159 |
0.54 |
| chr12_101026829_101027420 | 0.35 |
Ccdc88c |
coiled-coil domain containing 88C |
1785 |
0.23 |
| chr10_128400690_128401219 | 0.35 |
Slc39a5 |
solute carrier family 39 (metal ion transporter), member 5 |
256 |
0.73 |
| chr1_177411902_177412443 | 0.35 |
Gm26801 |
predicted gene, 26801 |
16912 |
0.17 |
| chr5_130022581_130023074 | 0.35 |
Asl |
argininosuccinate lyase |
1473 |
0.28 |
| chr11_99176689_99176840 | 0.35 |
Ccr7 |
chemokine (C-C motif) receptor 7 |
21687 |
0.13 |
| chr13_112043719_112044107 | 0.35 |
Gm15323 |
predicted gene 15323 |
38411 |
0.15 |
| chr2_26576075_26576641 | 0.34 |
Egfl7 |
EGF-like domain 7 |
3656 |
0.11 |
| chr7_46396141_46398423 | 0.34 |
Kcnc1 |
potassium voltage gated channel, Shaw-related subfamily, member 1 |
366 |
0.84 |
| chr4_155162564_155162715 | 0.34 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
25657 |
0.14 |
| chr2_153345876_153347096 | 0.34 |
2500004C02Rik |
RIKEN cDNA 2500004C02 gene |
413 |
0.6 |
| chr19_45230983_45235468 | 0.34 |
Lbx1 |
ladybird homeobox 1 |
2587 |
0.27 |
| chr11_96308497_96309471 | 0.34 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
2074 |
0.12 |
| chr4_62479845_62481162 | 0.33 |
Bspry |
B-box and SPRY domain containing |
414 |
0.75 |
| chr1_120598097_120600876 | 0.33 |
En1 |
engrailed 1 |
2932 |
0.29 |
| chr10_80879874_80880283 | 0.33 |
Tmprss9 |
transmembrane protease, serine 9 |
358 |
0.72 |
| chr2_70921160_70921456 | 0.33 |
Gm13632 |
predicted gene 13632 |
43479 |
0.14 |
| chr10_49779879_49781120 | 0.33 |
Grik2 |
glutamate receptor, ionotropic, kainate 2 (beta 2) |
2760 |
0.23 |
| chr3_51518589_51518921 | 0.33 |
Setd7 |
SET domain containing (lysine methyltransferase) 7 |
17892 |
0.09 |
| chr2_152085191_152085589 | 0.32 |
Scrt2 |
scratch family zinc finger 2 |
3861 |
0.17 |
| chr8_121590486_121591687 | 0.32 |
Map1lc3b |
microtubule-associated protein 1 light chain 3 beta |
374 |
0.77 |
| chr16_93366361_93366682 | 0.32 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
147 |
0.94 |
| chr7_30531338_30531929 | 0.32 |
Arhgap33 |
Rho GTPase activating protein 33 |
1768 |
0.12 |
| chr6_143832506_143833713 | 0.32 |
Sox5 |
SRY (sex determining region Y)-box 5 |
113979 |
0.06 |
| chr5_123222744_123223261 | 0.31 |
Psmd9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
5188 |
0.12 |
| chr12_16632574_16632725 | 0.31 |
Lpin1 |
lipin 1 |
14317 |
0.17 |
| chr7_19082814_19086200 | 0.31 |
Dmpk |
dystrophia myotonica-protein kinase |
203 |
0.83 |
| chr8_106336310_106337932 | 0.31 |
Smpd3 |
sphingomyelin phosphodiesterase 3, neutral |
867 |
0.61 |
| chr19_8709801_8709987 | 0.31 |
Slc3a2 |
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
3091 |
0.07 |
| chr11_83850990_83852424 | 0.30 |
Hnf1b |
HNF1 homeobox B |
535 |
0.69 |
| chr4_130171697_130173536 | 0.30 |
Tinagl1 |
tubulointerstitial nephritis antigen-like 1 |
2075 |
0.27 |
| chr1_89467693_89468170 | 0.30 |
Agap1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
12675 |
0.19 |
| chr3_146101204_146101488 | 0.29 |
Wdr63 |
WD repeat domain 63 |
6784 |
0.16 |
| chr4_140584429_140584695 | 0.29 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
219 |
0.94 |
| chr14_54408224_54410842 | 0.29 |
Slc7a7 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
73 |
0.94 |
| chr13_110094155_110094306 | 0.29 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
158918 |
0.04 |
| chr17_36019108_36019278 | 0.29 |
H2-T24 |
histocompatibility 2, T region locus 24 |
1332 |
0.17 |
| chr7_24369840_24369991 | 0.29 |
Kcnn4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
348 |
0.74 |
| chr9_41156579_41157745 | 0.28 |
Ubash3b |
ubiquitin associated and SH3 domain containing, B |
340 |
0.82 |
| chr6_136547141_136547342 | 0.28 |
Atf7ip |
activating transcription factor 7 interacting protein |
3946 |
0.19 |
| chr5_88564689_88565985 | 0.28 |
Rufy3 |
RUN and FYVE domain containing 3 |
297 |
0.87 |
| chr10_17651311_17651462 | 0.28 |
Gm47771 |
predicted gene, 47771 |
7127 |
0.25 |
| chrX_161719845_161720465 | 0.28 |
Rai2 |
retinoic acid induced 2 |
2528 |
0.41 |
| chr2_74692311_74693685 | 0.28 |
Hoxd10 |
homeobox D10 |
1074 |
0.19 |
| chr11_16503427_16503928 | 0.28 |
Sec61g |
SEC61, gamma subunit |
1367 |
0.46 |
| chr7_142474634_142476734 | 0.28 |
Lsp1 |
lymphocyte specific 1 |
690 |
0.52 |
| chr2_31638722_31641540 | 0.27 |
Prdm12 |
PR domain containing 12 |
94 |
0.84 |
| chr10_81145829_81146966 | 0.27 |
Zbtb7a |
zinc finger and BTB domain containing 7a |
8444 |
0.07 |
| chr14_32600761_32600912 | 0.27 |
Prrxl1 |
paired related homeobox protein-like 1 |
878 |
0.56 |
| chr2_158401974_158402125 | 0.27 |
Gm14201 |
predicted gene 14201 |
966 |
0.34 |
| chr15_8967409_8968940 | 0.27 |
Ranbp3l |
RAN binding protein 3-like |
172 |
0.96 |
| chr9_22049418_22049658 | 0.27 |
Elavl3 |
ELAV like RNA binding protein 3 |
2472 |
0.12 |
| chr11_84519041_84519440 | 0.27 |
Lhx1 |
LIM homeobox protein 1 |
2827 |
0.25 |
| chr2_66632905_66633478 | 0.27 |
Scn9a |
sodium channel, voltage-gated, type IX, alpha |
1462 |
0.53 |
| chr16_31754278_31755029 | 0.27 |
Dlg1 |
discs large MAGUK scaffold protein 1 |
303 |
0.86 |
| chr5_123201326_123201494 | 0.27 |
Gm43409 |
predicted gene 43409 |
9822 |
0.1 |
| chr1_152807871_152808731 | 0.26 |
Ncf2 |
neutrophil cytosolic factor 2 |
323 |
0.86 |
| chr11_61075424_61076210 | 0.26 |
Kcnj12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
10013 |
0.17 |
| chr17_28237484_28238264 | 0.26 |
Ppard |
peroxisome proliferator activator receptor delta |
5067 |
0.12 |
| chr3_89386729_89388779 | 0.26 |
Zbtb7b |
zinc finger and BTB domain containing 7B |
83 |
0.91 |
| chr7_35553510_35553693 | 0.26 |
Nudt19 |
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
1558 |
0.24 |
| chr2_155992156_155992465 | 0.26 |
Cep250 |
centrosomal protein 250 |
6395 |
0.11 |
| chr7_144238658_144240098 | 0.26 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
653 |
0.8 |
| chr2_174283563_174287177 | 0.26 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
12 |
0.53 |
| chr11_77892631_77894065 | 0.26 |
Pipox |
pipecolic acid oxidase |
748 |
0.58 |
| chr6_118757785_118758013 | 0.26 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
75 |
0.98 |
| chr14_70106795_70107014 | 0.25 |
Bin3 |
bridging integrator 3 |
6739 |
0.15 |
| chrX_48204804_48205976 | 0.25 |
Zdhhc9 |
zinc finger, DHHC domain containing 9 |
2938 |
0.23 |
| chr1_75212400_75213683 | 0.25 |
Stk16 |
serine/threonine kinase 16 |
20 |
0.92 |
| chr11_74555781_74555954 | 0.25 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
33116 |
0.15 |
| chr9_45202291_45202442 | 0.25 |
Tmprss4 |
transmembrane protease, serine 4 |
1620 |
0.23 |
| chr10_39736016_39736307 | 0.25 |
Rev3l |
REV3 like, DNA directed polymerase zeta catalytic subunit |
3797 |
0.16 |
| chr7_25152042_25153378 | 0.25 |
D930028M14Rik |
RIKEN cDNA D930028M14 gene |
253 |
0.87 |
| chr1_130731543_130732516 | 0.25 |
AA986860 |
expressed sequence AA986860 |
53 |
0.94 |
| chr18_74733826_74734119 | 0.25 |
Myo5b |
myosin VB |
207 |
0.94 |
| chr6_60827248_60827682 | 0.25 |
Snca |
synuclein, alpha |
91 |
0.97 |
| chr2_25459014_25459284 | 0.25 |
Paxx |
non-homologous end joining factor |
1051 |
0.23 |
| chr11_104234777_104235157 | 0.25 |
Mapt |
microtubule-associated protein tau |
3383 |
0.19 |
| chr5_64159427_64160996 | 0.25 |
Tbc1d1 |
TBC1 domain family, member 1 |
0 |
0.97 |
| chr17_78290191_78290620 | 0.24 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
10222 |
0.18 |
| chr4_119027493_119027644 | 0.24 |
Gm12862 |
predicted gene 12862 |
27083 |
0.08 |
| chr18_82489298_82489862 | 0.24 |
Mbp |
myelin basic protein |
14210 |
0.17 |
| chr4_125488664_125489133 | 0.24 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
1802 |
0.36 |
| chr17_49436615_49437040 | 0.24 |
Mocs1 |
molybdenum cofactor synthesis 1 |
3631 |
0.26 |
| chr18_83192582_83193531 | 0.24 |
1700095A13Rik |
RIKEN cDNA 1700095A13 gene |
85166 |
0.08 |
| chr15_80190031_80190182 | 0.24 |
Mgat3 |
mannoside acetylglucosaminyltransferase 3 |
16385 |
0.11 |
| chr7_24370279_24371432 | 0.24 |
Kcnn4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
517 |
0.59 |
| chr1_167375050_167375216 | 0.24 |
Aldh9a1 |
aldehyde dehydrogenase 9, subfamily A1 |
10967 |
0.13 |
| chr12_4873247_4874779 | 0.24 |
Mfsd2b |
major facilitator superfamily domain containing 2B |
332 |
0.82 |
| chr3_96576165_96577539 | 0.24 |
Gm16253 |
predicted gene 16253 |
132 |
0.89 |
| chr2_158308324_158308475 | 0.24 |
Lbp |
lipopolysaccharide binding protein |
1787 |
0.24 |
| chr6_113282134_113283787 | 0.24 |
Cpne9 |
copine family member IX |
595 |
0.55 |
| chr15_102981032_102982455 | 0.24 |
Hoxc9 |
homeobox C9 |
4711 |
0.09 |
| chr3_87913699_87914234 | 0.23 |
Hdgf |
heparin binding growth factor |
5 |
0.95 |
| chr12_102355566_102356003 | 0.23 |
Rin3 |
Ras and Rab interactor 3 |
1004 |
0.59 |
| chr14_34623313_34624132 | 0.23 |
Opn4 |
opsin 4 (melanopsin) |
23580 |
0.11 |
| chr9_120481695_120481846 | 0.23 |
Eif1b |
eukaryotic translation initiation factor 1B |
10462 |
0.14 |
| chr14_56100230_56100381 | 0.23 |
Ctsg |
cathepsin G |
1470 |
0.21 |
| chr19_4058692_4059651 | 0.23 |
Gstp3 |
glutathione S-transferase pi 3 |
124 |
0.88 |
| chr16_18418352_18418503 | 0.23 |
Gm15764 |
predicted gene 15764 |
1542 |
0.24 |
| chr4_140244024_140245271 | 0.23 |
Igsf21 |
immunoglobulin superfamily, member 21 |
2137 |
0.36 |
| chr3_89138382_89138543 | 0.23 |
Pklr |
pyruvate kinase liver and red blood cell |
1839 |
0.14 |
| chr4_46391513_46391664 | 0.23 |
Trmo |
tRNA methyltransferase O |
2151 |
0.22 |
| chr2_181767278_181768191 | 0.23 |
Myt1 |
myelin transcription factor 1 |
222 |
0.91 |
| chr12_13180179_13180570 | 0.23 |
Gm24208 |
predicted gene, 24208 |
16611 |
0.16 |
| chr2_20736484_20738247 | 0.23 |
Etl4 |
enhancer trap locus 4 |
51 |
0.98 |
| chr1_136445067_136445535 | 0.23 |
Ddx59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
12899 |
0.14 |
| chr10_12963484_12964773 | 0.23 |
Stx11 |
syntaxin 11 |
130 |
0.96 |
| chr11_117946846_117947224 | 0.23 |
Socs3 |
suppressor of cytokine signaling 3 |
22151 |
0.11 |
| chr3_57426561_57426712 | 0.22 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
1322 |
0.5 |
| chr16_30387562_30389582 | 0.22 |
Atp13a3 |
ATPase type 13A3 |
42 |
0.98 |
| chrX_137118132_137120673 | 0.22 |
Esx1 |
extraembryonic, spermatogenesis, homeobox 1 |
769 |
0.36 |
| chr11_97511052_97512791 | 0.22 |
Gm11611 |
predicted gene 11611 |
9974 |
0.12 |
| chr1_132365874_132366736 | 0.22 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
767 |
0.54 |
| chr1_194676054_194676333 | 0.22 |
Gm38036 |
predicted gene, 38036 |
809 |
0.58 |
| chr1_42707054_42709031 | 0.22 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
10 |
0.97 |
| chr18_23807443_23808685 | 0.22 |
Mapre2 |
microtubule-associated protein, RP/EB family, member 2 |
3961 |
0.2 |
| chr10_42533045_42533196 | 0.22 |
Snx3 |
sorting nexin 3 |
30830 |
0.14 |
| chr5_140614430_140614581 | 0.22 |
Lfng |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
2820 |
0.18 |
| chr16_97658836_97658987 | 0.22 |
Tmprss2 |
transmembrane protease, serine 2 |
47716 |
0.14 |
| chr10_44011393_44011544 | 0.22 |
Crybg1 |
crystallin beta-gamma domain containing 1 |
7099 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.2 | 0.6 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.2 | 0.6 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.3 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.3 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.1 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.3 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.2 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.5 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.1 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.3 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.2 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.2 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 0.2 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.8 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.3 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.3 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.0 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 1.0 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.2 | GO:0002591 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0010513 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) regulation of phosphatidylinositol biosynthetic process(GO:0010511) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002923) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.4 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.8 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.4 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0034784 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0052717 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |