Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
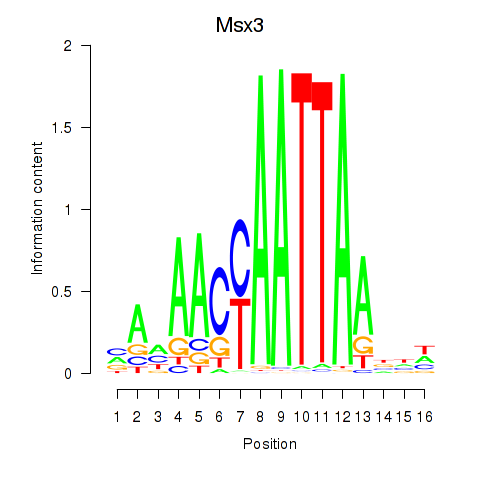
Results for Msx3
Z-value: 0.90
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSMUSG00000025469.9 | msh homeobox 3 |
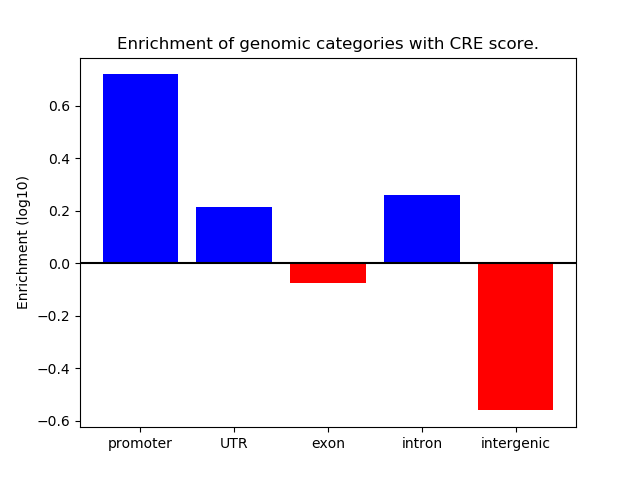
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_140060622_140060773 | Msx3 | 11608 | 0.084238 | -0.27 | 3.8e-02 | Click! |
| chr7_140048576_140049822 | Msx3 | 110 | 0.924623 | 0.25 | 5.1e-02 | Click! |
| chr7_140047755_140048553 | Msx3 | 934 | 0.360646 | 0.21 | 1.1e-01 | Click! |
| chr7_140046752_140046903 | Msx3 | 2261 | 0.149192 | -0.18 | 1.8e-01 | Click! |
| chr7_140047010_140047161 | Msx3 | 2003 | 0.165125 | -0.18 | 1.8e-01 | Click! |
Activity of the Msx3 motif across conditions
Conditions sorted by the z-value of the Msx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
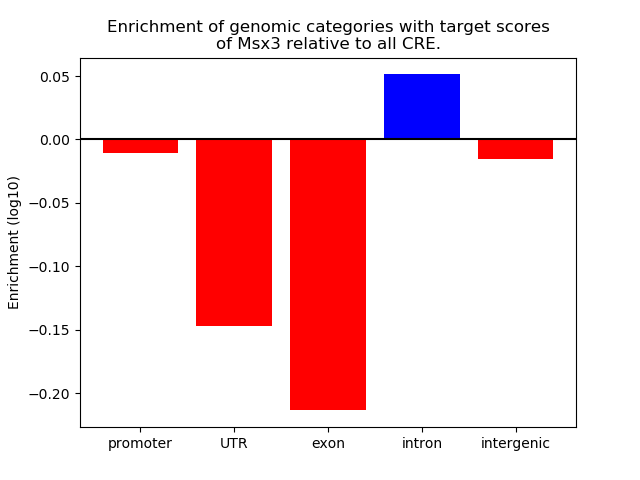
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_33083500_33083959 | 3.85 |
Trdn |
triadin |
168 |
0.97 |
| chr8_85361454_85361605 | 2.65 |
Mylk3 |
myosin light chain kinase 3 |
3795 |
0.16 |
| chr15_40656769_40657228 | 2.48 |
Zfpm2 |
zinc finger protein, multitype 2 |
1647 |
0.51 |
| chr9_79976568_79977294 | 2.42 |
Filip1 |
filamin A interacting protein 1 |
728 |
0.66 |
| chr1_153330166_153330548 | 2.33 |
Lamc1 |
laminin, gamma 1 |
2429 |
0.26 |
| chr10_3368375_3368998 | 2.33 |
Ppp1r14c |
protein phosphatase 1, regulatory inhibitor subunit 14C |
2142 |
0.36 |
| chr18_43436329_43436751 | 2.14 |
Dpysl3 |
dihydropyrimidinase-like 3 |
1746 |
0.35 |
| chrX_164437518_164437860 | 2.12 |
Asb11 |
ankyrin repeat and SOCS box-containing 11 |
369 |
0.85 |
| chr1_187999444_187999905 | 2.06 |
Esrrg |
estrogen-related receptor gamma |
1806 |
0.42 |
| chr10_30838379_30838546 | 2.04 |
Hey2 |
hairy/enhancer-of-split related with YRPW motif 2 |
3553 |
0.21 |
| chr5_75975434_75975824 | 2.02 |
Kdr |
kinase insert domain protein receptor |
2829 |
0.25 |
| chr14_63235525_63236294 | 1.99 |
Gata4 |
GATA binding protein 4 |
9339 |
0.17 |
| chr1_140182332_140182724 | 1.93 |
Cfh |
complement component factor h |
752 |
0.73 |
| chr7_109191327_109192334 | 1.92 |
Lmo1 |
LIM domain only 1 |
16623 |
0.17 |
| chr5_138609015_138609166 | 1.81 |
Gm15497 |
predicted gene 15497 |
1649 |
0.2 |
| chr15_85673548_85673944 | 1.79 |
Lncppara |
long noncoding RNA near Ppara |
20130 |
0.13 |
| chr1_40806618_40806769 | 1.78 |
Tmem182 |
transmembrane protein 182 |
1092 |
0.48 |
| chrX_138915472_138915880 | 1.76 |
Nrk |
Nik related kinase |
1245 |
0.61 |
| chr3_19646421_19646748 | 1.66 |
Trim55 |
tripartite motif-containing 55 |
2076 |
0.26 |
| chr14_59478693_59479244 | 1.56 |
Cab39l |
calcium binding protein 39-like |
152 |
0.96 |
| chr2_143959771_143959922 | 1.55 |
Rrbp1 |
ribosome binding protein 1 |
2201 |
0.29 |
| chr2_45001616_45002114 | 1.53 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
21050 |
0.19 |
| chr10_128178900_128179051 | 1.51 |
Rbms2 |
RNA binding motif, single stranded interacting protein 2 |
1322 |
0.23 |
| chr13_29628489_29628949 | 1.48 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
111265 |
0.07 |
| chr16_43503039_43503193 | 1.47 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
498 |
0.85 |
| chr16_43504464_43505047 | 1.45 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1058 |
0.61 |
| chr5_75975825_75977229 | 1.45 |
Kdr |
kinase insert domain protein receptor |
1931 |
0.31 |
| chr1_169745785_169746010 | 1.45 |
Rgs4 |
regulator of G-protein signaling 4 |
1726 |
0.41 |
| chr14_57034631_57034809 | 1.43 |
B020004C17Rik |
RIKEN cDNA B020004C17 gene |
19586 |
0.15 |
| chr10_23686915_23687430 | 1.42 |
4930520K02Rik |
RIKEN cDNA 4930520K02 gene |
65024 |
0.09 |
| chr5_57945633_57945798 | 1.41 |
Gm42639 |
predicted gene 42639 |
673 |
0.56 |
| chr13_46423537_46423785 | 1.39 |
Rbm24 |
RNA binding motif protein 24 |
1839 |
0.41 |
| chr13_46420548_46420948 | 1.39 |
Rbm24 |
RNA binding motif protein 24 |
197 |
0.96 |
| chr5_138608490_138608879 | 1.32 |
Zfp68 |
zinc finger protein 68 |
1627 |
0.2 |
| chr2_14825138_14825659 | 1.31 |
Cacnb2 |
calcium channel, voltage-dependent, beta 2 subunit |
1306 |
0.4 |
| chr13_83566010_83566411 | 1.29 |
Mef2c |
myocyte enhancer factor 2C |
6138 |
0.29 |
| chr10_39165752_39166209 | 1.29 |
Ccn6 |
cellular communication network factor 6 |
2186 |
0.23 |
| chr3_30009457_30010152 | 1.28 |
Mecom |
MDS1 and EVI1 complex locus |
1635 |
0.36 |
| chr6_98525524_98525718 | 1.26 |
4930595L18Rik |
RIKEN cDNA 4930595L18 gene |
26486 |
0.26 |
| chr2_20942943_20943436 | 1.26 |
Arhgap21 |
Rho GTPase activating protein 21 |
230 |
0.93 |
| chr8_106985656_106985807 | 1.26 |
Gm22085 |
predicted gene, 22085 |
23880 |
0.1 |
| chr12_60595918_60596185 | 1.24 |
Gm6068 |
predicted gene 6068 |
47813 |
0.16 |
| chr16_96205810_96206739 | 1.20 |
Sh3bgr |
SH3-binding domain glutamic acid-rich protein |
369 |
0.83 |
| chr18_37694128_37695315 | 1.20 |
Pcdhga5 |
protocadherin gamma subfamily A, 5 |
341 |
0.59 |
| chr16_77502046_77502387 | 1.19 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
1832 |
0.24 |
| chr13_73262153_73264451 | 1.18 |
Irx4 |
Iroquois homeobox 4 |
2805 |
0.22 |
| chr14_91522550_91522701 | 1.17 |
Gm48942 |
predicted gene, 48942 |
125214 |
0.06 |
| chr3_65635958_65636417 | 1.17 |
Gm38233 |
predicted gene, 38233 |
20013 |
0.12 |
| chr10_93307247_93307878 | 1.16 |
Elk3 |
ELK3, member of ETS oncogene family |
3249 |
0.21 |
| chr6_14753955_14754106 | 1.16 |
Ppp1r3a |
protein phosphatase 1, regulatory subunit 3A |
1244 |
0.63 |
| chr13_71961770_71962476 | 1.15 |
Irx1 |
Iroquois homeobox 1 |
1407 |
0.5 |
| chr15_40167052_40167633 | 1.15 |
Gm33301 |
predicted gene, 33301 |
4735 |
0.21 |
| chr5_102727171_102727322 | 1.13 |
Arhgap24 |
Rho GTPase activating protein 24 |
2273 |
0.45 |
| chr3_16818309_16818538 | 1.13 |
Gm26485 |
predicted gene, 26485 |
4889 |
0.36 |
| chr2_55439535_55439810 | 1.12 |
Kcnj3 |
potassium inwardly-rectifying channel, subfamily J, member 3 |
2507 |
0.37 |
| chr16_28898704_28899105 | 1.11 |
Mb21d2 |
Mab-21 domain containing 2 |
30769 |
0.21 |
| chr14_49793886_49794107 | 1.10 |
Gm22989 |
predicted gene, 22989 |
5712 |
0.15 |
| chr9_115096411_115096891 | 1.08 |
Osbpl10 |
oxysterol binding protein-like 10 |
7810 |
0.23 |
| chr3_60530332_60531570 | 1.07 |
Mbnl1 |
muscleblind like splicing factor 1 |
1320 |
0.48 |
| chr4_40851422_40852119 | 1.06 |
Gm25931 |
predicted gene, 25931 |
1368 |
0.21 |
| chr13_83529350_83529841 | 1.06 |
Mef2c |
myocyte enhancer factor 2C |
543 |
0.86 |
| chr10_96744086_96744687 | 1.06 |
Gm6859 |
predicted gene 6859 |
25442 |
0.15 |
| chr1_107590023_107591215 | 1.05 |
Serpinb8 |
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
573 |
0.79 |
| chr5_72796212_72796844 | 1.05 |
Tec |
tec protein tyrosine kinase |
10508 |
0.15 |
| chr12_34529024_34529175 | 1.04 |
Hdac9 |
histone deacetylase 9 |
225 |
0.96 |
| chr5_116456914_116457389 | 1.04 |
Gm42853 |
predicted gene 42853 |
333 |
0.83 |
| chr15_102997344_102997495 | 1.03 |
Hoxc6 |
homeobox C6 |
838 |
0.37 |
| chr18_11060814_11061287 | 1.03 |
Gata6 |
GATA binding protein 6 |
2003 |
0.37 |
| chr1_177199086_177199237 | 1.02 |
Gm37706 |
predicted gene, 37706 |
28299 |
0.14 |
| chr1_81593421_81594539 | 1.00 |
Gm6198 |
predicted gene 6198 |
36497 |
0.2 |
| chr12_66935099_66935250 | 1.00 |
Gm47989 |
predicted gene, 47989 |
7237 |
0.29 |
| chr6_146500226_146500471 | 1.00 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
1530 |
0.4 |
| chr10_39612918_39613668 | 0.99 |
Traf3ip2 |
TRAF3 interacting protein 2 |
359 |
0.48 |
| chr12_31264979_31266396 | 0.99 |
Lamb1 |
laminin B1 |
390 |
0.78 |
| chr9_83664457_83665200 | 0.98 |
Gm36120 |
predicted gene, 36120 |
22697 |
0.17 |
| chr8_57315271_57316273 | 0.98 |
Hand2os1 |
Hand2, opposite strand 1 |
62 |
0.96 |
| chr5_147722674_147723044 | 0.98 |
Flt1 |
FMS-like tyrosine kinase 1 |
3129 |
0.27 |
| chr15_18282470_18282621 | 0.97 |
4921515E04Rik |
RIKEN cDNA 4921515E04 gene |
22745 |
0.24 |
| chr1_188002104_188003122 | 0.97 |
Esrrg |
estrogen-related receptor gamma |
4745 |
0.26 |
| chr2_107292125_107293014 | 0.97 |
Kcna4 |
potassium voltage-gated channel, shaker-related subfamily, member 4 |
1174 |
0.64 |
| chr9_86438288_86439112 | 0.97 |
Ube2cbp |
ubiquitin-conjugating enzyme E2C binding protein |
26250 |
0.17 |
| chr10_71028206_71028357 | 0.96 |
Gm48025 |
predicted gene, 48025 |
35241 |
0.14 |
| chr10_128173680_128173831 | 0.96 |
Rbms2 |
RNA binding motif, single stranded interacting protein 2 |
2724 |
0.12 |
| chr18_75372210_75374418 | 0.96 |
Smad7 |
SMAD family member 7 |
1600 |
0.41 |
| chr3_79180505_79180656 | 0.95 |
Rapgef2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
760 |
0.66 |
| chr10_30845489_30845963 | 0.95 |
Hey2 |
hairy/enhancer-of-split related with YRPW motif 2 |
2925 |
0.24 |
| chr18_60375901_60376327 | 0.93 |
Iigp1 |
interferon inducible GTPase 1 |
67 |
0.96 |
| chr3_79144294_79146166 | 0.93 |
Rapgef2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
253 |
0.94 |
| chr10_115306134_115306562 | 0.93 |
Rab21 |
RAB21, member RAS oncogene family |
9243 |
0.15 |
| chr14_119602272_119602445 | 0.93 |
Gm6212 |
predicted gene 6212 |
41654 |
0.2 |
| chr5_97387372_97387523 | 0.92 |
Naa11 |
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
4932 |
0.21 |
| chr17_81723922_81724212 | 0.91 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
14310 |
0.27 |
| chr9_77344708_77344864 | 0.91 |
Mlip |
muscular LMNA-interacting protein |
3007 |
0.22 |
| chr3_9573681_9573832 | 0.90 |
Zfp704 |
zinc finger protein 704 |
8457 |
0.25 |
| chr15_100635478_100635706 | 0.90 |
Smagp |
small cell adhesion glycoprotein |
33 |
0.94 |
| chr7_48842956_48843247 | 0.89 |
Csrp3 |
cysteine and glycine-rich protein 3 |
550 |
0.71 |
| chr9_67261068_67261449 | 0.89 |
Tln2 |
talin 2 |
835 |
0.67 |
| chr2_104467722_104467873 | 0.89 |
Hipk3 |
homeodomain interacting protein kinase 3 |
25893 |
0.14 |
| chr12_72441609_72442616 | 0.88 |
Lrrc9 |
leucine rich repeat containing 9 |
77 |
0.97 |
| chr5_5261065_5262223 | 0.88 |
Cdk14 |
cyclin-dependent kinase 14 |
3665 |
0.25 |
| chr10_58394969_58395625 | 0.88 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
909 |
0.6 |
| chr3_76077829_76078163 | 0.86 |
Fstl5 |
follistatin-like 5 |
2413 |
0.35 |
| chr10_22815720_22817074 | 0.86 |
Gm10824 |
predicted gene 10824 |
648 |
0.66 |
| chr15_50870065_50870337 | 0.86 |
Trps1 |
transcriptional repressor GATA binding 1 |
12605 |
0.21 |
| chrY_3871382_3871533 | 0.86 |
Gm18177 |
predicted gene, 18177 |
4471 |
0.13 |
| chr4_116073957_116074118 | 0.85 |
Uqcrh |
ubiquinol-cytochrome c reductase hinge protein |
994 |
0.31 |
| chr4_112748048_112748199 | 0.85 |
Skint10 |
selection and upkeep of intraepithelial T cells 10 |
26702 |
0.2 |
| chr18_55375824_55376307 | 0.84 |
Gm37828 |
predicted gene, 37828 |
36553 |
0.21 |
| chr16_85807463_85807781 | 0.84 |
Adamts1 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
4509 |
0.26 |
| chr18_60439220_60439471 | 0.84 |
BC023105 |
cDNA sequence BC023105 |
2420 |
0.24 |
| chr1_140180086_140180415 | 0.83 |
Cfh |
complement component factor h |
3030 |
0.32 |
| chr10_84968503_84968659 | 0.83 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31188 |
0.19 |
| chr17_17448568_17449744 | 0.82 |
Lix1 |
limb and CNS expressed 1 |
336 |
0.88 |
| chr6_87495688_87496317 | 0.82 |
Arhgap25 |
Rho GTPase activating protein 25 |
312 |
0.88 |
| chr7_98678348_98678798 | 0.81 |
Gm45132 |
predicted gene 45132 |
2074 |
0.24 |
| chr1_23321078_23322043 | 0.81 |
Gm20954 |
predicted gene, 20954 |
25039 |
0.11 |
| chr10_99263341_99263492 | 0.81 |
Dusp6 |
dual specificity phosphatase 6 |
185 |
0.9 |
| chr10_110922562_110923038 | 0.80 |
Csrp2 |
cysteine and glycine-rich protein 2 |
2595 |
0.22 |
| chr3_9947586_9947797 | 0.80 |
Gm17877 |
predicted gene, 17877 |
24310 |
0.16 |
| chr3_116966712_116967946 | 0.80 |
4930455H04Rik |
RIKEN cDNA 4930455H04 gene |
938 |
0.41 |
| chr5_124192601_124192752 | 0.80 |
Pitpnm2os2 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 2 |
2231 |
0.19 |
| chr10_45338438_45338763 | 0.80 |
Bves |
blood vessel epicardial substance |
2828 |
0.24 |
| chr4_137863933_137864084 | 0.80 |
Ece1 |
endothelin converting enzyme 1 |
1711 |
0.38 |
| chr3_78594754_78595081 | 0.80 |
Gm15442 |
predicted gene 15442 |
106280 |
0.07 |
| chr13_9494460_9495468 | 0.79 |
Gm48871 |
predicted gene, 48871 |
49064 |
0.12 |
| chr9_73967774_73969221 | 0.79 |
Unc13c |
unc-13 homolog C |
469 |
0.88 |
| chr1_34439512_34439753 | 0.79 |
Ccdc115 |
coiled-coil domain containing 115 |
40 |
0.71 |
| chr13_102957440_102958679 | 0.78 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
287 |
0.95 |
| chr6_39055880_39056082 | 0.78 |
1700025N23Rik |
RIKEN cDNA 1700025N23 gene |
11617 |
0.15 |
| chr16_37779366_37779517 | 0.78 |
Fstl1 |
follistatin-like 1 |
2289 |
0.31 |
| chr14_66147812_66148016 | 0.77 |
Chrna2 |
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
6981 |
0.17 |
| chr5_14026773_14026948 | 0.77 |
Gm43519 |
predicted gene 43519 |
1034 |
0.45 |
| chr15_51854353_51854609 | 0.77 |
Eif3h |
eukaryotic translation initiation factor 3, subunit H |
10973 |
0.17 |
| chr2_67568406_67568614 | 0.77 |
B3galt1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
2639 |
0.3 |
| chr7_136275874_136276030 | 0.77 |
C030029H02Rik |
RIKEN cDNA C030029H02 gene |
7618 |
0.21 |
| chr18_47370463_47370942 | 0.77 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
1832 |
0.37 |
| chr8_125186745_125187429 | 0.77 |
Gm16237 |
predicted gene 16237 |
40236 |
0.17 |
| chr6_29351906_29352057 | 0.76 |
Calu |
calumenin |
3862 |
0.13 |
| chr8_45901486_45902198 | 0.76 |
Pdlim3 |
PDZ and LIM domain 3 |
13190 |
0.12 |
| chr1_162000010_162000265 | 0.76 |
Dnm3 |
dynamin 3 |
4123 |
0.16 |
| chr13_71967461_71968595 | 0.76 |
Irx1 |
Iroquois homeobox 1 |
4305 |
0.28 |
| chr4_31468474_31468780 | 0.75 |
Gm11922 |
predicted gene 11922 |
88155 |
0.1 |
| chr6_16316653_16317675 | 0.75 |
Gm3148 |
predicted gene 3148 |
79537 |
0.1 |
| chrX_13208320_13208567 | 0.75 |
Rpl3-ps1 |
ribosomal protein L3, pseudogene 1 |
5872 |
0.11 |
| chr10_56106927_56107500 | 0.75 |
Msl3l2 |
MSL3 like 2 |
296 |
0.92 |
| chr17_88597076_88597227 | 0.75 |
Ston1 |
stonin 1 |
533 |
0.74 |
| chr6_94592745_94593176 | 0.75 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
12433 |
0.18 |
| chr1_168429421_168430005 | 0.74 |
Pbx1 |
pre B cell leukemia homeobox 1 |
1791 |
0.44 |
| chr13_104948155_104948442 | 0.73 |
Gm4938 |
predicted pseudogene 4938 |
29518 |
0.2 |
| chr14_28533395_28533564 | 0.73 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
22028 |
0.19 |
| chrX_159943977_159944189 | 0.72 |
Sh3kbp1 |
SH3-domain kinase binding protein 1 |
1065 |
0.59 |
| chr18_24711980_24712131 | 0.72 |
Fhod3 |
formin homology 2 domain containing 3 |
2610 |
0.23 |
| chr5_16630062_16630213 | 0.72 |
Gm42975 |
predicted gene 42975 |
24415 |
0.2 |
| chrX_83936299_83936450 | 0.72 |
Dmd |
dystrophin, muscular dystrophy |
8474 |
0.32 |
| chr3_42111234_42111590 | 0.72 |
Gm37846 |
predicted gene, 37846 |
52453 |
0.16 |
| chr13_52528312_52528571 | 0.72 |
Diras2 |
DIRAS family, GTP-binding RAS-like 2 |
2838 |
0.37 |
| chrX_142684293_142684706 | 0.71 |
Tmem164 |
transmembrane protein 164 |
1611 |
0.4 |
| chr9_53850790_53851038 | 0.71 |
Sln |
sarcolipin |
750 |
0.65 |
| chr9_56416072_56416694 | 0.71 |
Gm36940 |
predicted gene, 36940 |
317 |
0.81 |
| chr13_95765042_95765602 | 0.71 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
925 |
0.57 |
| chr1_131232374_131233673 | 0.70 |
Rassf5 |
Ras association (RalGDS/AF-6) domain family member 5 |
552 |
0.67 |
| chr18_61783794_61784202 | 0.70 |
Afap1l1 |
actin filament associated protein 1-like 1 |
2613 |
0.25 |
| chr8_13491946_13492269 | 0.70 |
Gas6 |
growth arrest specific 6 |
2383 |
0.25 |
| chr1_65186243_65186463 | 0.70 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
85 |
0.79 |
| chr19_31764456_31765203 | 0.70 |
Prkg1 |
protein kinase, cGMP-dependent, type I |
144 |
0.98 |
| chr10_14017752_14018957 | 0.70 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
51330 |
0.13 |
| chr12_91551959_91552341 | 0.69 |
Tshr |
thyroid stimulating hormone receptor |
14084 |
0.16 |
| chr11_118744160_118745307 | 0.69 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
16308 |
0.21 |
| chr3_96562001_96562194 | 0.69 |
Txnip |
thioredoxin interacting protein |
2043 |
0.13 |
| chr10_20776929_20777274 | 0.68 |
Gm48651 |
predicted gene, 48651 |
20984 |
0.21 |
| chr15_97765080_97766053 | 0.68 |
Rapgef3os2 |
Rap guanine nucleotide exchange factor (GEF) 3, opposite strand 2 |
983 |
0.36 |
| chr8_34994050_34994306 | 0.68 |
Gm34368 |
predicted gene, 34368 |
14304 |
0.16 |
| chr5_20583581_20583870 | 0.68 |
Gm3544 |
predicted gene 3544 |
4871 |
0.26 |
| chr7_35948219_35949093 | 0.67 |
Gm28514 |
predicted gene 28514 |
110396 |
0.06 |
| chr9_60742965_60743393 | 0.67 |
1700036A12Rik |
RIKEN cDNA 1700036A12 gene |
59 |
0.97 |
| chr3_142390298_142390537 | 0.67 |
Pdlim5 |
PDZ and LIM domain 5 |
1391 |
0.52 |
| chr11_110358024_110358368 | 0.67 |
Abca5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
20480 |
0.22 |
| chr17_63865820_63865971 | 0.67 |
Fer |
fer (fms/fps related) protein kinase |
1880 |
0.34 |
| chr1_62795476_62796303 | 0.67 |
Gm37121 |
predicted gene, 37121 |
13413 |
0.19 |
| chr11_65266805_65267363 | 0.67 |
Myocd |
myocardin |
2770 |
0.29 |
| chr7_61309034_61309202 | 0.67 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
2595 |
0.4 |
| chr11_70018020_70018411 | 0.67 |
Dlg4 |
discs large MAGUK scaffold protein 4 |
226 |
0.8 |
| chr15_57659816_57660144 | 0.67 |
Zhx2 |
zinc fingers and homeoboxes 2 |
34685 |
0.17 |
| chrX_166344270_166344507 | 0.66 |
Gpm6b |
glycoprotein m6b |
304 |
0.92 |
| chr8_35592634_35593539 | 0.66 |
Gm16793 |
predicted gene, 16793 |
4114 |
0.21 |
| chr16_59599420_59599980 | 0.65 |
Crybg3 |
beta-gamma crystallin domain containing 3 |
1279 |
0.47 |
| chr13_110396048_110396669 | 0.65 |
Plk2 |
polo like kinase 2 |
346 |
0.91 |
| chr19_45855235_45855499 | 0.65 |
Gm50196 |
predicted gene, 50196 |
11750 |
0.15 |
| chr1_90204802_90205464 | 0.65 |
Ackr3 |
atypical chemokine receptor 3 |
1127 |
0.47 |
| chr14_118080663_118081120 | 0.65 |
Dct |
dopachrome tautomerase |
28647 |
0.15 |
| chr5_105052746_105052954 | 0.65 |
Gbp8 |
guanylate-binding protein 8 |
706 |
0.66 |
| chr10_103015192_103015343 | 0.65 |
Alx1 |
ALX homeobox 1 |
7307 |
0.21 |
| chr2_70315394_70315699 | 0.64 |
Rpl9-ps7 |
ribosomal protein L9, pseudogene 7 |
143436 |
0.04 |
| chr11_31875409_31875592 | 0.64 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
2225 |
0.33 |
| chr17_81480235_81480868 | 0.64 |
Gm11096 |
predicted gene 11096 |
38788 |
0.2 |
| chr11_120186805_120187341 | 0.64 |
2810410L24Rik |
RIKEN cDNA 2810410L24 gene |
1678 |
0.23 |
| chr2_57997628_57998772 | 0.63 |
Galnt5 |
polypeptide N-acetylgalactosaminyltransferase 5 |
316 |
0.9 |
| chr17_10293681_10293832 | 0.63 |
Qk |
quaking |
10823 |
0.25 |
| chr6_118194255_118194406 | 0.63 |
Ret |
ret proto-oncogene |
2803 |
0.25 |
| chr9_95597786_95598558 | 0.62 |
Gm28164 |
predicted gene 28164 |
24247 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.6 | 1.9 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.6 | 2.4 | GO:0003195 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.6 | 1.7 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.5 | 1.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.3 | 1.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 0.6 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 0.7 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.2 | 1.4 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.4 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.2 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.7 | GO:1900378 | regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 0.5 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.2 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.5 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.4 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.7 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 1.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.6 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.7 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.3 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 1.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 | 0.3 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.1 | 0.3 | GO:0035907 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.5 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 | 0.3 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 2.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.1 | 0.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.2 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.3 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.1 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.2 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 1.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.2 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.1 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.1 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0060399 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.6 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 1.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:1900200 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.0 | GO:0072298 | regulation of metanephric glomerulus development(GO:0072298) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0043379 | memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 1.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.2 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.0 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.1 | GO:0071295 | cellular response to vitamin(GO:0071295) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:1901859 | regulation of mitochondrial DNA replication(GO:0090296) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0048369 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.0 | GO:1901723 | negative regulation of cell proliferation involved in kidney development(GO:1901723) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:1901201 | regulation of extracellular matrix assembly(GO:1901201) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.0 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0051187 | cofactor catabolic process(GO:0051187) |
| 0.0 | 0.0 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.1 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 1.2 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 1.4 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.3 | 1.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.3 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 1.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.2 | 1.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 0.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.4 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.1 | 1.6 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.2 | GO:0015368 | calcium:sodium antiporter activity(GO:0005432) calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0043955 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 4.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0034821 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 2.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0044688 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0022821 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 3.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0016937 | short-branched-chain-acyl-CoA dehydrogenase activity(GO:0016937) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.0 | GO:0034786 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0044466 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |