Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
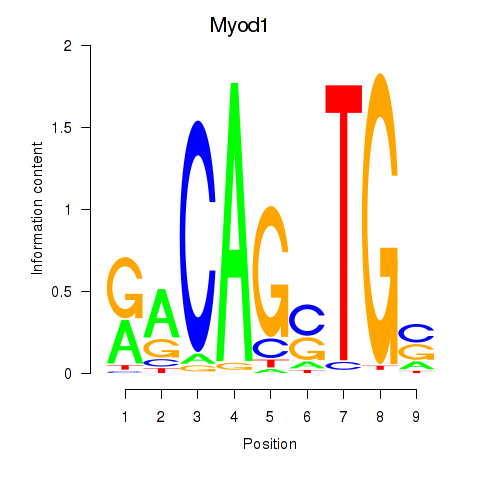
Results for Myod1
Z-value: 0.52
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSMUSG00000009471.3 | myogenic differentiation 1 |
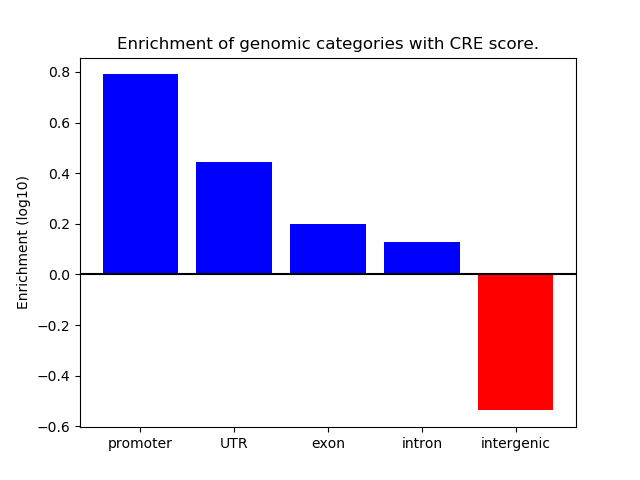
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_46376321_46379126 | Myod1 | 1249 | 0.401431 | 0.28 | 2.9e-02 | Click! |
| chr7_46375692_46376219 | Myod1 | 519 | 0.741117 | 0.18 | 1.6e-01 | Click! |
| chr7_46375296_46375447 | Myod1 | 1103 | 0.446948 | 0.08 | 5.5e-01 | Click! |
Activity of the Myod1 motif across conditions
Conditions sorted by the z-value of the Myod1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
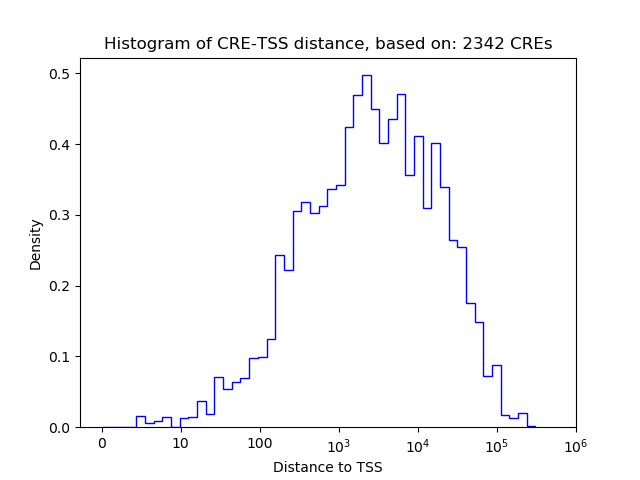
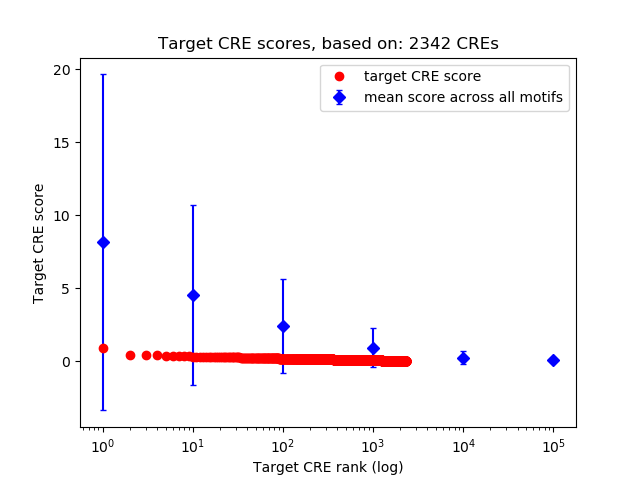
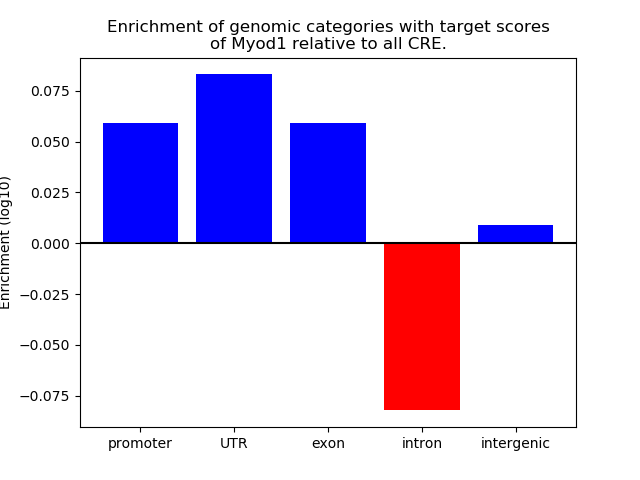
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_162217585_162219370 | 0.94 |
Dnm3os |
dynamin 3, opposite strand |
601 |
0.46 |
| chr16_38363598_38364152 | 0.42 |
Popdc2 |
popeye domain containing 2 |
1630 |
0.26 |
| chr9_115096411_115096891 | 0.42 |
Osbpl10 |
oxysterol binding protein-like 10 |
7810 |
0.23 |
| chr8_57326741_57329467 | 0.40 |
Hand2os1 |
Hand2, opposite strand 1 |
3871 |
0.15 |
| chr4_11385045_11386187 | 0.38 |
Esrp1 |
epithelial splicing regulatory protein 1 |
778 |
0.6 |
| chr13_55835316_55837389 | 0.37 |
Pitx1 |
paired-like homeodomain transcription factor 1 |
160 |
0.62 |
| chr18_65348348_65348930 | 0.37 |
Gm41757 |
predicted gene, 41757 |
518 |
0.56 |
| chr3_128990459_128991248 | 0.35 |
Gm9387 |
predicted pseudogene 9387 |
35755 |
0.19 |
| chr12_73039135_73040425 | 0.33 |
Six1 |
sine oculis-related homeobox 1 |
3035 |
0.27 |
| chr9_20972154_20973375 | 0.33 |
S1pr2 |
sphingosine-1-phosphate receptor 2 |
1214 |
0.26 |
| chr13_63273798_63275383 | 0.32 |
Gm47585 |
predicted gene, 47585 |
990 |
0.28 |
| chr12_112747021_112747471 | 0.32 |
Cep170b |
centrosomal protein 170B |
3282 |
0.12 |
| chr18_65083730_65084940 | 0.31 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
1281 |
0.54 |
| chr8_87938260_87939386 | 0.31 |
Zfp423 |
zinc finger protein 423 |
5229 |
0.29 |
| chr1_135324343_135325907 | 0.31 |
Lmod1 |
leiomodin 1 (smooth muscle) |
318 |
0.83 |
| chr9_87729583_87731038 | 0.31 |
Tbx18 |
T-box18 |
187 |
0.9 |
| chr17_68002299_68002969 | 0.31 |
Arhgap28 |
Rho GTPase activating protein 28 |
1486 |
0.54 |
| chr7_44474127_44475010 | 0.31 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
1030 |
0.25 |
| chr12_9573836_9576858 | 0.30 |
Osr1 |
odd-skipped related transcription factor 1 |
906 |
0.56 |
| chr2_180389757_180390438 | 0.29 |
Mir1a-1 |
microRNA 1a-1 |
1049 |
0.41 |
| chr16_44015370_44016774 | 0.29 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr10_39614434_39615372 | 0.29 |
Gm16364 |
predicted gene 16364 |
1221 |
0.36 |
| chr9_120021835_120022273 | 0.29 |
Xirp1 |
xin actin-binding repeat containing 1 |
1544 |
0.23 |
| chr12_4523121_4523682 | 0.29 |
Gm31938 |
predicted gene, 31938 |
712 |
0.6 |
| chr9_21851806_21852608 | 0.28 |
Dock6 |
dedicator of cytokinesis 6 |
409 |
0.74 |
| chr11_47377104_47378325 | 0.27 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
1808 |
0.52 |
| chr11_85885788_85887022 | 0.27 |
Tbx4 |
T-box 4 |
17 |
0.97 |
| chr11_100144835_100146198 | 0.27 |
Krt19 |
keratin 19 |
604 |
0.52 |
| chr1_106629982_106630261 | 0.27 |
Gm37053 |
predicted gene, 37053 |
23251 |
0.19 |
| chr5_113798574_113798890 | 0.27 |
Tmem119 |
transmembrane protein 119 |
1714 |
0.21 |
| chr1_14303973_14305599 | 0.26 |
Eya1 |
EYA transcriptional coactivator and phosphatase 1 |
158 |
0.97 |
| chr7_6727103_6728704 | 0.26 |
Peg3 |
paternally expressed 3 |
2516 |
0.14 |
| chr2_35623294_35623923 | 0.26 |
Dab2ip |
disabled 2 interacting protein |
1448 |
0.46 |
| chr8_107545084_107547731 | 0.26 |
Wwp2 |
WW domain containing E3 ubiquitin protein ligase 2 |
1395 |
0.4 |
| chr16_21207155_21207444 | 0.26 |
Ephb3 |
Eph receptor B3 |
2544 |
0.25 |
| chr6_24599678_24600100 | 0.25 |
Lmod2 |
leiomodin 2 (cardiac) |
2127 |
0.25 |
| chr4_43520861_43521689 | 0.25 |
Tpm2 |
tropomyosin 2, beta |
1606 |
0.17 |
| chr14_101886064_101886474 | 0.25 |
Lmo7 |
LIM domain only 7 |
2150 |
0.43 |
| chr11_69714656_69715350 | 0.25 |
Gm12307 |
predicted gene 12307 |
15581 |
0.05 |
| chr8_121118797_121121438 | 0.24 |
Foxc2 |
forkhead box C2 |
3946 |
0.14 |
| chr17_42741899_42742151 | 0.24 |
Adgrf2 |
adhesion G protein-coupled receptor F2 |
154 |
0.96 |
| chr17_44301052_44301539 | 0.24 |
Clic5 |
chloride intracellular channel 5 |
64189 |
0.12 |
| chr9_92276388_92276986 | 0.23 |
Plscr2 |
phospholipid scramblase 2 |
941 |
0.48 |
| chr3_105815846_105816287 | 0.23 |
Gm5547 |
predicted gene 5547 |
499 |
0.69 |
| chr7_114103782_114104353 | 0.23 |
Rras2 |
related RAS viral (r-ras) oncogene 2 |
13697 |
0.21 |
| chr3_146117173_146118635 | 0.23 |
Mcoln3 |
mucolipin 3 |
454 |
0.73 |
| chr9_66512511_66514532 | 0.23 |
Fbxl22 |
F-box and leucine-rich repeat protein 22 |
1088 |
0.47 |
| chr13_55825087_55826711 | 0.23 |
Gm47071 |
predicted gene, 47071 |
351 |
0.82 |
| chr17_24752448_24753648 | 0.23 |
Hs3st6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
32 |
0.93 |
| chr4_43399011_43400247 | 0.23 |
Rusc2 |
RUN and SH3 domain containing 2 |
1609 |
0.28 |
| chr15_101293230_101294713 | 0.23 |
Smim41 |
small integral membrane protein 41 |
739 |
0.47 |
| chr11_114216624_114216941 | 0.22 |
1700092K14Rik |
RIKEN cDNA 1700092K14 gene |
18524 |
0.15 |
| chr1_75360071_75361415 | 0.22 |
Des |
desmin |
414 |
0.72 |
| chr14_34585878_34586349 | 0.22 |
Ldb3 |
LIM domain binding 3 |
2368 |
0.19 |
| chr2_163694015_163694536 | 0.22 |
Pkig |
protein kinase inhibitor, gamma |
237 |
0.89 |
| chr11_115974294_115975742 | 0.22 |
Itgb4 |
integrin beta 4 |
39 |
0.95 |
| chr4_114907257_114907947 | 0.22 |
Foxd2 |
forkhead box D2 |
1271 |
0.3 |
| chr6_51193157_51193734 | 0.22 |
Mir148a |
microRNA 148a |
76465 |
0.1 |
| chr13_72629730_72632045 | 0.22 |
Irx2 |
Iroquois homeobox 2 |
1062 |
0.51 |
| chr8_41015082_41015919 | 0.21 |
Mtus1 |
mitochondrial tumor suppressor 1 |
820 |
0.53 |
| chr17_88791510_88792237 | 0.21 |
Lhcgr |
luteinizing hormone/choriogonadotropin receptor |
98 |
0.98 |
| chr9_74110075_74111065 | 0.21 |
Wdr72 |
WD repeat domain 72 |
166 |
0.97 |
| chr19_55746711_55749175 | 0.21 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
5098 |
0.32 |
| chr9_56712934_56713139 | 0.21 |
Lingo1 |
leucine rich repeat and Ig domain containing 1 |
27783 |
0.18 |
| chr3_87951379_87952516 | 0.21 |
Crabp2 |
cellular retinoic acid binding protein II |
197 |
0.87 |
| chr4_141302731_141302975 | 0.21 |
Epha2 |
Eph receptor A2 |
446 |
0.71 |
| chr8_111745500_111746511 | 0.21 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
2196 |
0.3 |
| chr4_134237503_134238650 | 0.20 |
Cnksr1 |
connector enhancer of kinase suppressor of Ras 1 |
263 |
0.8 |
| chr7_140899782_140900558 | 0.20 |
Cox8b |
cytochrome c oxidase subunit 8B |
276 |
0.74 |
| chr18_29242584_29242819 | 0.20 |
Gm34743 |
predicted gene, 34743 |
2646 |
0.43 |
| chr11_8622429_8622770 | 0.20 |
Tns3 |
tensin 3 |
2288 |
0.45 |
| chr2_173057598_173057795 | 0.20 |
Gm14453 |
predicted gene 14453 |
23116 |
0.12 |
| chr5_137610176_137611504 | 0.20 |
Pcolce |
procollagen C-endopeptidase enhancer protein |
138 |
0.86 |
| chr5_30913519_30915234 | 0.20 |
Emilin1 |
elastin microfibril interfacer 1 |
637 |
0.48 |
| chr13_31625167_31627270 | 0.20 |
Gm27516 |
predicted gene, 27516 |
325 |
0.4 |
| chr11_102623866_102624954 | 0.20 |
2810433D01Rik |
RIKEN cDNA 2810433D01 gene |
6 |
0.95 |
| chr9_44136029_44136582 | 0.20 |
Mcam |
melanoma cell adhesion molecule |
239 |
0.78 |
| chr18_76542618_76543294 | 0.20 |
Gm31933 |
predicted gene, 31933 |
89914 |
0.09 |
| chr2_70083482_70084035 | 0.20 |
Myo3b |
myosin IIIB |
684 |
0.74 |
| chr16_26041199_26042258 | 0.20 |
AU015336 |
expressed sequence AU015336 |
460 |
0.86 |
| chr13_102903771_102904870 | 0.20 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
1466 |
0.56 |
| chr4_124276838_124278353 | 0.20 |
Gm37667 |
predicted gene, 37667 |
1582 |
0.35 |
| chr2_69673818_69673969 | 0.19 |
Klhl41 |
kelch-like 41 |
3773 |
0.18 |
| chr3_138069501_138070764 | 0.19 |
1110002E22Rik |
RIKEN cDNA 1110002E22 gene |
1465 |
0.3 |
| chr7_17061715_17062460 | 0.19 |
Hif3a |
hypoxia inducible factor 3, alpha subunit |
297 |
0.82 |
| chr9_45203520_45204009 | 0.19 |
Tmprss4 |
transmembrane protease, serine 4 |
222 |
0.87 |
| chr2_84423814_84425562 | 0.19 |
Calcrl |
calcitonin receptor-like |
578 |
0.75 |
| chr7_18957439_18958650 | 0.19 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
29356 |
0.06 |
| chr1_74035856_74036007 | 0.19 |
Tns1 |
tensin 1 |
1202 |
0.53 |
| chr19_25411022_25411281 | 0.19 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
4452 |
0.24 |
| chr10_79988072_79989720 | 0.19 |
Cnn2 |
calponin 2 |
273 |
0.76 |
| chr19_24861287_24862270 | 0.19 |
Pgm5 |
phosphoglucomutase 5 |
77 |
0.96 |
| chr1_51291546_51292409 | 0.19 |
Cavin2 |
caveolae associated 2 |
2851 |
0.27 |
| chr12_33966605_33968831 | 0.19 |
Twist1 |
twist basic helix-loop-helix transcription factor 1 |
10047 |
0.22 |
| chr11_96334278_96335532 | 0.19 |
Hoxb3 |
homeobox B3 |
6040 |
0.08 |
| chr4_124701261_124701890 | 0.19 |
Fhl3 |
four and a half LIM domains 3 |
855 |
0.37 |
| chr8_80119316_80119579 | 0.19 |
Gm5909 |
predicted gene 5909 |
7361 |
0.19 |
| chr2_28138697_28138848 | 0.18 |
F730016J06Rik |
RIKEN cDNA F730016J06 gene |
20565 |
0.17 |
| chr14_67271890_67272768 | 0.18 |
Gm23178 |
predicted gene, 23178 |
2634 |
0.24 |
| chr9_24874511_24875131 | 0.18 |
Amd-ps6 |
S-adenosylmethionine decarboxylase, pseudogene 6 |
47476 |
0.12 |
| chr2_35627849_35628114 | 0.18 |
Dab2ip |
disabled 2 interacting protein |
5821 |
0.24 |
| chr7_112228733_112229227 | 0.18 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3090 |
0.35 |
| chr1_40874622_40874941 | 0.18 |
Gm5973 |
predicted gene 5973 |
10549 |
0.23 |
| chr15_89425017_89425997 | 0.18 |
Cpt1b |
carnitine palmitoyltransferase 1b, muscle |
288 |
0.74 |
| chr13_92425754_92427030 | 0.18 |
Ankrd34b |
ankyrin repeat domain 34B |
51 |
0.94 |
| chr1_88639699_88640131 | 0.18 |
Gm38130 |
predicted gene, 38130 |
13170 |
0.17 |
| chr4_148960341_148961414 | 0.18 |
Gm17029 |
predicted gene 17029 |
517 |
0.74 |
| chr9_21311433_21311747 | 0.18 |
Ap1m2 |
adaptor protein complex AP-1, mu 2 subunit |
721 |
0.46 |
| chr7_70280148_70281631 | 0.18 |
Gm29327 |
predicted gene 29327 |
31113 |
0.13 |
| chr3_133233594_133234317 | 0.18 |
Arhgef38 |
Rho guanine nucleotide exchange factor (GEF) 38 |
934 |
0.64 |
| chr3_68585286_68585461 | 0.18 |
Schip1 |
schwannomin interacting protein 1 |
1194 |
0.53 |
| chr9_59947366_59947592 | 0.18 |
Nr2e3 |
nuclear receptor subfamily 2, group E, member 3 |
1312 |
0.32 |
| chr11_113611912_113612498 | 0.18 |
Sstr2 |
somatostatin receptor 2 |
7137 |
0.18 |
| chr3_8736196_8736672 | 0.18 |
C230057A21Rik |
RIKEN cDNA C230057A21 gene |
74 |
0.97 |
| chr6_145942273_145942684 | 0.18 |
Gm43931 |
predicted gene, 43931 |
4598 |
0.18 |
| chr8_71684084_71684803 | 0.18 |
Insl3 |
insulin-like 3 |
4771 |
0.09 |
| chr18_60963369_60963520 | 0.18 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
110 |
0.95 |
| chr3_30104992_30105810 | 0.17 |
Mecom |
MDS1 and EVI1 complex locus |
35022 |
0.17 |
| chr6_14903005_14903973 | 0.17 |
Foxp2 |
forkhead box P2 |
32 |
0.99 |
| chr14_63236760_63237176 | 0.17 |
Gata4 |
GATA binding protein 4 |
8280 |
0.17 |
| chr9_71770829_71771315 | 0.17 |
Cgnl1 |
cingulin-like 1 |
488 |
0.81 |
| chr3_135285527_135285798 | 0.17 |
Bdh2 |
3-hydroxybutyrate dehydrogenase, type 2 |
2996 |
0.21 |
| chr17_62655730_62656448 | 0.17 |
Gm25800 |
predicted gene, 25800 |
198969 |
0.03 |
| chr2_93177441_93177592 | 0.17 |
Trp53i11 |
transformation related protein 53 inducible protein 11 |
10032 |
0.22 |
| chr7_49522042_49523415 | 0.17 |
Nav2 |
neuron navigator 2 |
25464 |
0.21 |
| chr6_120275123_120275344 | 0.17 |
B4galnt3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
19326 |
0.19 |
| chr5_119842721_119843952 | 0.17 |
Tbx5 |
T-box 5 |
7181 |
0.18 |
| chr1_18961494_18962019 | 0.17 |
Gm24075 |
predicted gene, 24075 |
8077 |
0.26 |
| chr16_87553258_87554074 | 0.17 |
Map3k7cl |
Map3k7 C-terminal like |
336 |
0.84 |
| chr7_121735353_121735504 | 0.17 |
Scnn1g |
sodium channel, nonvoltage-gated 1 gamma |
949 |
0.56 |
| chr16_38360437_38360588 | 0.17 |
Popdc2 |
popeye domain containing 2 |
1697 |
0.25 |
| chr3_19622651_19622802 | 0.17 |
1700064H15Rik |
RIKEN cDNA 1700064H15 gene |
5951 |
0.17 |
| chr16_78326082_78326665 | 0.17 |
Cxadr |
coxsackie virus and adenovirus receptor |
1166 |
0.43 |
| chr16_42152536_42153083 | 0.17 |
Lsamp |
limbic system-associated membrane protein |
8784 |
0.22 |
| chr8_18740237_18741640 | 0.17 |
Angpt2 |
angiopoietin 2 |
624 |
0.75 |
| chr10_75174015_75174616 | 0.17 |
Bcr |
BCR activator of RhoGEF and GTPase |
350 |
0.89 |
| chr11_55414788_55415644 | 0.17 |
Sparc |
secreted acidic cysteine rich glycoprotein |
4682 |
0.18 |
| chr6_54272742_54273928 | 0.17 |
Chn2 |
chimerin 2 |
427 |
0.82 |
| chr2_30850400_30850833 | 0.17 |
Prrx2 |
paired related homeobox 2 |
5172 |
0.14 |
| chr1_75362633_75362784 | 0.17 |
Des |
desmin |
678 |
0.52 |
| chr7_141117549_141118467 | 0.17 |
Ano9 |
anoctamin 9 |
202 |
0.85 |
| chr3_69548831_69549124 | 0.17 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
2 |
0.98 |
| chr5_113136296_113136937 | 0.17 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
1370 |
0.27 |
| chr11_106365272_106365607 | 0.17 |
Prr29 |
proline rich 29 |
33 |
0.96 |
| chr16_18696451_18696829 | 0.16 |
Rps2-ps7 |
ribosomal protein S2, pseudogene 7 |
14424 |
0.15 |
| chr1_189924428_189924893 | 0.16 |
Smyd2 |
SET and MYND domain containing 2 |
2297 |
0.28 |
| chr5_81519288_81520108 | 0.16 |
Gm43084 |
predicted gene 43084 |
16028 |
0.29 |
| chr17_12744783_12745031 | 0.16 |
Airn |
antisense Igf2r RNA |
3509 |
0.17 |
| chr15_73707761_73707912 | 0.16 |
Gpr20 |
G protein-coupled receptor 20 |
331 |
0.86 |
| chr4_57295053_57295204 | 0.16 |
Gm12536 |
predicted gene 12536 |
4968 |
0.2 |
| chr9_40325276_40326082 | 0.16 |
1700110K17Rik |
RIKEN cDNA 1700110K17 gene |
2253 |
0.19 |
| chr8_26272856_26273881 | 0.16 |
Gm31727 |
predicted gene, 31727 |
6391 |
0.14 |
| chr1_93190721_93191081 | 0.16 |
Crocc2 |
ciliary rootlet coiled-coil, rootletin family member 2 |
22176 |
0.11 |
| chr8_113433901_113434458 | 0.16 |
Gm25766 |
predicted gene, 25766 |
85427 |
0.09 |
| chr13_23497689_23499269 | 0.16 |
Btn2a2 |
butyrophilin, subfamily 2, member A2 |
9622 |
0.06 |
| chr6_139586816_139587911 | 0.16 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
126 |
0.96 |
| chr7_100926314_100930096 | 0.16 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
3902 |
0.17 |
| chr2_84634714_84636973 | 0.16 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
2262 |
0.17 |
| chr3_131110318_131112630 | 0.16 |
Lef1os1 |
LEF1 opposite strand RNA 1 |
616 |
0.56 |
| chr9_107550029_107550198 | 0.16 |
Zmynd10 |
zinc finger, MYND domain containing 10 |
376 |
0.48 |
| chr15_96135146_96135802 | 0.16 |
D030018L15Rik |
RIKEN cDNA D030018L15 gene |
55921 |
0.13 |
| chr13_46428917_46429384 | 0.16 |
Rbm24 |
RNA binding motif protein 24 |
7328 |
0.24 |
| chr12_38778127_38778471 | 0.16 |
Etv1 |
ets variant 1 |
1081 |
0.56 |
| chr3_55779960_55781204 | 0.16 |
Mab21l1 |
mab-21-like 1 |
1928 |
0.32 |
| chr3_57576670_57576821 | 0.16 |
Wwtr1 |
WW domain containing transcription regulator 1 |
835 |
0.56 |
| chr18_66344941_66345611 | 0.16 |
Ccbe1 |
collagen and calcium binding EGF domains 1 |
42535 |
0.1 |
| chr13_60455009_60455742 | 0.16 |
Gm35333 |
predicted gene, 35333 |
20644 |
0.15 |
| chr1_189775009_189775805 | 0.16 |
Ptpn14 |
protein tyrosine phosphatase, non-receptor type 14 |
11229 |
0.19 |
| chr7_19461513_19461793 | 0.16 |
Gm23099 |
predicted gene, 23099 |
718 |
0.38 |
| chr1_58713328_58714310 | 0.16 |
Cflar |
CASP8 and FADD-like apoptosis regulator |
67 |
0.96 |
| chr8_121086245_121087227 | 0.16 |
Gm27530 |
predicted gene, 27530 |
2030 |
0.19 |
| chr3_51778939_51779197 | 0.16 |
Gm37342 |
predicted gene, 37342 |
16980 |
0.11 |
| chr4_148780321_148780472 | 0.16 |
Casz1 |
castor zinc finger 1 |
24033 |
0.18 |
| chr15_101139270_101139421 | 0.16 |
Acvrl1 |
activin A receptor, type II-like 1 |
1745 |
0.23 |
| chr12_23188694_23189971 | 0.16 |
Gm47385 |
predicted gene, 47385 |
66 |
0.96 |
| chr7_28070851_28071079 | 0.15 |
Fcgbp |
Fc fragment of IgG binding protein |
271 |
0.86 |
| chr2_13577101_13577578 | 0.15 |
Vim |
vimentin |
2030 |
0.37 |
| chr8_25343082_25343881 | 0.15 |
5430421F17Rik |
RIKEN cDNA 5430421F17 gene |
37211 |
0.12 |
| chr5_38549575_38549794 | 0.15 |
Gm22249 |
predicted gene, 22249 |
3226 |
0.2 |
| chr10_79639823_79639974 | 0.15 |
Shc2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
1980 |
0.14 |
| chr12_26068051_26068784 | 0.15 |
Gm47750 |
predicted gene, 47750 |
81899 |
0.09 |
| chr10_94520068_94521446 | 0.15 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
5900 |
0.23 |
| chr2_13579434_13579672 | 0.15 |
Vim |
vimentin |
184 |
0.96 |
| chr10_88336673_88337098 | 0.15 |
Dram1 |
DNA-damage regulated autophagy modulator 1 |
20111 |
0.14 |
| chr5_127227734_127228696 | 0.15 |
Tmem132c |
transmembrane protein 132C |
13593 |
0.18 |
| chr5_123059978_123060693 | 0.15 |
Gm6444 |
predicted gene 6444 |
5907 |
0.09 |
| chr9_14675747_14675965 | 0.15 |
Gm19790 |
predicted gene, 19790 |
4114 |
0.11 |
| chr6_89345563_89345754 | 0.15 |
Gm44207 |
predicted gene, 44207 |
518 |
0.74 |
| chr2_132944858_132946039 | 0.15 |
Fermt1 |
fermitin family member 1 |
181 |
0.93 |
| chr19_28504036_28504436 | 0.15 |
Glis3 |
GLIS family zinc finger 3 |
36184 |
0.2 |
| chr16_85797660_85797940 | 0.15 |
Adamts1 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 |
1838 |
0.41 |
| chr16_32486028_32486482 | 0.15 |
Slc51a |
solute carrier family 51, alpha subunit |
1448 |
0.3 |
| chr6_56617179_56617515 | 0.15 |
Gm22493 |
predicted gene, 22493 |
16398 |
0.17 |
| chr5_111221096_111221247 | 0.15 |
Ttc28 |
tetratricopeptide repeat domain 28 |
4287 |
0.22 |
| chr9_44085798_44086457 | 0.15 |
Usp2 |
ubiquitin specific peptidase 2 |
1063 |
0.25 |
| chr17_34497017_34497248 | 0.15 |
Btnl5-ps |
butyrophilin-like 5, pseudogene |
297 |
0.75 |
| chr18_50087652_50087803 | 0.15 |
C030005K06Rik |
RIKEN cDNA C030005K06 gene |
33883 |
0.13 |
| chr18_35739288_35740804 | 0.15 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
30 |
0.95 |
| chr5_113335226_113336256 | 0.15 |
Mir7230 |
microRNA 7230 |
1904 |
0.22 |
| chr10_24958395_24959363 | 0.15 |
Gm36172 |
predicted gene, 36172 |
31260 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.1 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.2 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.4 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0048369 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.3 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.0 | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis(GO:1902338) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.0 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.0 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.0 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0018641 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |