Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
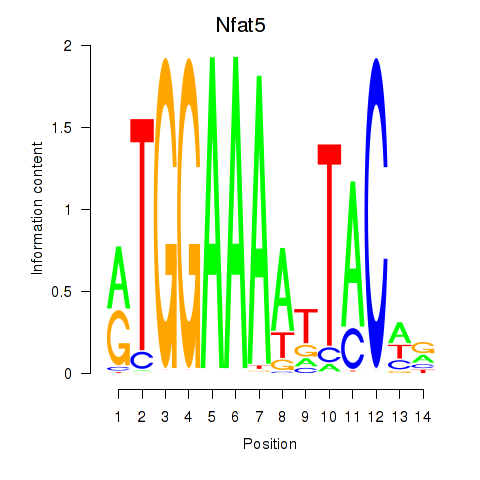
Results for Nfat5
Z-value: 0.79
Transcription factors associated with Nfat5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfat5
|
ENSMUSG00000003847.10 | nuclear factor of activated T cells 5 |
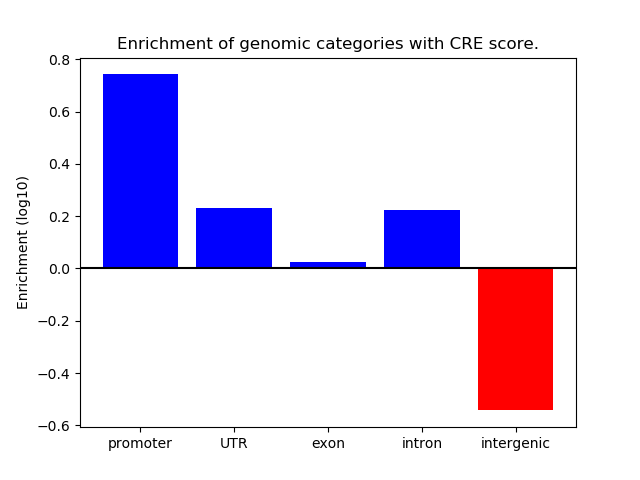
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_107336573_107336890 | Nfat5 | 1777 | 0.363267 | 0.30 | 1.8e-02 | Click! |
| chr8_107297033_107297184 | Nfat5 | 3549 | 0.242841 | 0.26 | 4.1e-02 | Click! |
| chr8_107344888_107345039 | Nfat5 | 177 | 0.951000 | -0.24 | 6.1e-02 | Click! |
| chr8_107295537_107295867 | Nfat5 | 2143 | 0.317928 | 0.23 | 7.9e-02 | Click! |
| chr8_107292893_107293194 | Nfat5 | 427 | 0.843177 | 0.10 | 4.3e-01 | Click! |
Activity of the Nfat5 motif across conditions
Conditions sorted by the z-value of the Nfat5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
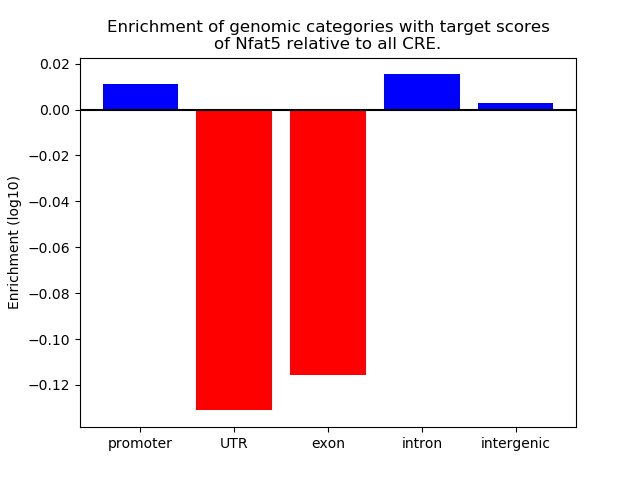
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_139586816_139587911 | 1.58 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
126 |
0.96 |
| chr3_19647545_19647696 | 1.53 |
Trim55 |
tripartite motif-containing 55 |
3112 |
0.2 |
| chr1_164455819_164456603 | 1.45 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1811 |
0.27 |
| chr17_81734587_81734955 | 1.41 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
3606 |
0.34 |
| chr17_72921491_72924008 | 1.37 |
Lbh |
limb-bud and heart |
1561 |
0.47 |
| chr2_35691764_35692937 | 1.35 |
Dab2ip |
disabled 2 interacting protein |
17 |
0.98 |
| chr14_59478693_59479244 | 1.33 |
Cab39l |
calcium binding protein 39-like |
152 |
0.96 |
| chr1_130732649_130733832 | 1.24 |
AA986860 |
expressed sequence AA986860 |
1130 |
0.29 |
| chr14_31166307_31168596 | 1.18 |
Stab1 |
stabilin 1 |
1144 |
0.34 |
| chr18_4350648_4350799 | 1.14 |
Map3k8 |
mitogen-activated protein kinase kinase kinase 8 |
2234 |
0.19 |
| chr18_12256920_12257106 | 1.14 |
Ankrd29 |
ankyrin repeat domain 29 |
14684 |
0.14 |
| chrX_74248072_74248576 | 1.13 |
Flna |
filamin, alpha |
1496 |
0.18 |
| chr8_28593556_28594184 | 1.10 |
Gm26795 |
predicted gene, 26795 |
338 |
0.92 |
| chr13_106835269_106836682 | 1.10 |
Lrrc70 |
leucine rich repeat containing 70 |
173 |
0.7 |
| chr16_74900702_74901266 | 1.08 |
Gm49674 |
predicted gene, 49674 |
25046 |
0.23 |
| chr10_18447141_18447345 | 1.02 |
Nhsl1 |
NHS-like 1 |
22645 |
0.22 |
| chr17_43389439_43390317 | 1.00 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
390 |
0.89 |
| chr6_115982816_115983479 | 0.99 |
Plxnd1 |
plexin D1 |
11858 |
0.14 |
| chr2_148856545_148856768 | 0.99 |
Cstdc2 |
cystatin domain containing 2 |
5722 |
0.13 |
| chr1_164454554_164455430 | 0.94 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1215 |
0.39 |
| chr5_32140229_32140654 | 0.93 |
Fosl2 |
fos-like antigen 2 |
4268 |
0.18 |
| chr5_39716146_39716373 | 0.92 |
Hs3st1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
39216 |
0.18 |
| chr3_153783659_153783895 | 0.91 |
Gm10042 |
predicted gene 10042 |
1276 |
0.34 |
| chr7_112217172_112217348 | 0.91 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
8596 |
0.27 |
| chr7_130576739_130577783 | 0.89 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
177 |
0.94 |
| chr16_50752363_50752789 | 0.89 |
Dubr |
Dppa2 upstream binding RNA |
19803 |
0.17 |
| chr11_83849571_83850989 | 0.89 |
Hnf1b |
HNF1 homeobox B |
217 |
0.83 |
| chr8_78771411_78771855 | 0.87 |
Gm47209 |
predicted gene, 47209 |
2912 |
0.24 |
| chr6_17309182_17310034 | 0.87 |
Cav1 |
caveolin 1, caveolae protein |
1885 |
0.33 |
| chr6_119052982_119053231 | 0.86 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
26901 |
0.24 |
| chr5_29477932_29478829 | 0.86 |
Mnx1 |
motor neuron and pancreas homeobox 1 |
90 |
0.96 |
| chr18_76200378_76201192 | 0.86 |
Gm9028 |
predicted gene 9028 |
23398 |
0.15 |
| chrX_82949777_82950054 | 0.85 |
Dmd |
dystrophin, muscular dystrophy |
1012 |
0.71 |
| chr15_8967409_8968940 | 0.84 |
Ranbp3l |
RAN binding protein 3-like |
172 |
0.96 |
| chr7_75655315_75655603 | 0.83 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
11274 |
0.18 |
| chr17_81736193_81736564 | 0.82 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
1999 |
0.44 |
| chr9_105520039_105521535 | 0.82 |
Atp2c1 |
ATPase, Ca++-sequestering |
360 |
0.86 |
| chr14_63247276_63249044 | 0.81 |
Gata4 |
GATA binding protein 4 |
2889 |
0.24 |
| chr19_27364312_27364550 | 0.81 |
Gm50113 |
predicted gene, 50113 |
1641 |
0.36 |
| chr8_127499907_127500504 | 0.81 |
Pard3 |
par-3 family cell polarity regulator |
52459 |
0.15 |
| chr3_79885722_79887545 | 0.80 |
Gm36569 |
predicted gene, 36569 |
173 |
0.83 |
| chr18_45269269_45269613 | 0.80 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
436 |
0.85 |
| chr14_20663037_20663711 | 0.80 |
Synpo2l |
synaptopodin 2-like |
1200 |
0.26 |
| chr14_120296391_120298017 | 0.79 |
Mbnl2 |
muscleblind like splicing factor 2 |
470 |
0.87 |
| chr14_54959527_54959749 | 0.78 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
1731 |
0.13 |
| chr1_144055657_144055889 | 0.78 |
Rgs2 |
regulator of G-protein signaling 2 |
51612 |
0.16 |
| chr5_116419135_116419697 | 0.78 |
Hspb8 |
heat shock protein 8 |
3448 |
0.16 |
| chr6_57533615_57533966 | 0.78 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
1636 |
0.32 |
| chr6_124919073_124920498 | 0.77 |
Ptms |
parathymosin |
318 |
0.73 |
| chr11_107335368_107335799 | 0.77 |
Pitpnc1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
1943 |
0.23 |
| chr8_69887012_69889642 | 0.76 |
Yjefn3 |
YjeF N-terminal domain containing 3 |
142 |
0.84 |
| chr1_138608112_138608354 | 0.76 |
Nek7 |
NIMA (never in mitosis gene a)-related expressed kinase 7 |
11441 |
0.18 |
| chr3_66292264_66292972 | 0.76 |
Veph1 |
ventricular zone expressed PH domain-containing 1 |
4130 |
0.27 |
| chr16_97606744_97607108 | 0.75 |
Tmprss2 |
transmembrane protease, serine 2 |
4043 |
0.25 |
| chr19_57275523_57275688 | 0.74 |
4930449E18Rik |
RIKEN cDNA 4930449E18 gene |
1645 |
0.38 |
| chr5_141414827_141414978 | 0.74 |
Gm16035 |
predicted gene 16035 |
33976 |
0.16 |
| chr6_37431738_37432032 | 0.74 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
10261 |
0.27 |
| chr13_97390464_97390915 | 0.74 |
Gm33447 |
predicted gene, 33447 |
14455 |
0.18 |
| chr8_80811787_80812072 | 0.73 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
18955 |
0.18 |
| chr8_111923307_111923610 | 0.73 |
Tmem231 |
transmembrane protein 231 |
4702 |
0.15 |
| chr5_143558813_143558964 | 0.73 |
Fam220a |
family with sequence similarity 220, member A |
9848 |
0.14 |
| chr1_23271255_23272383 | 0.72 |
Mir30a |
microRNA 30a |
450 |
0.73 |
| chr3_151749374_151749990 | 0.72 |
Ifi44 |
interferon-induced protein 44 |
200 |
0.95 |
| chr12_73587091_73587345 | 0.72 |
Prkch |
protein kinase C, eta |
2375 |
0.29 |
| chr9_56866808_56868850 | 0.71 |
Cspg4 |
chondroitin sulfate proteoglycan 4 |
2796 |
0.18 |
| chr7_126624812_126625701 | 0.69 |
Nupr1 |
nuclear protein transcription regulator 1 |
88 |
0.92 |
| chr3_153689877_153690164 | 0.69 |
Gm22206 |
predicted gene, 22206 |
18891 |
0.16 |
| chr7_137526118_137526299 | 0.69 |
Gm45397 |
predicted gene 45397 |
38380 |
0.18 |
| chr1_188003839_188004226 | 0.69 |
9330162B11Rik |
RIKEN cDNA 9330162B11 gene |
4958 |
0.26 |
| chr15_68937001_68937382 | 0.69 |
Khdrbs3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7123 |
0.23 |
| chr16_93938522_93938959 | 0.67 |
Cldn14 |
claudin 14 |
3644 |
0.18 |
| chr1_23261578_23261729 | 0.67 |
Gm6420 |
predicted gene 6420 |
5246 |
0.13 |
| chr16_23611686_23611850 | 0.67 |
Rtp4 |
receptor transporter protein 4 |
1843 |
0.39 |
| chrX_71964243_71964394 | 0.67 |
Prrg3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
1336 |
0.41 |
| chr5_119841347_119842608 | 0.67 |
Tbx5 |
T-box 5 |
5822 |
0.19 |
| chr13_89541543_89541900 | 0.66 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
1925 |
0.42 |
| chr11_96254039_96254881 | 0.66 |
Gm53 |
predicted gene 53 |
1928 |
0.15 |
| chr13_108497868_108498217 | 0.66 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
48033 |
0.17 |
| chr6_134151167_134151791 | 0.66 |
Gm17088 |
predicted gene 17088 |
20528 |
0.18 |
| chr17_50678196_50678377 | 0.66 |
Gm7334 |
predicted gene 7334 |
20402 |
0.25 |
| chr8_14996081_14996572 | 0.66 |
Arhgef10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
190 |
0.91 |
| chr10_98624940_98625347 | 0.66 |
Gm5427 |
predicted gene 5427 |
74567 |
0.11 |
| chr5_65157791_65158129 | 0.65 |
Klhl5 |
kelch-like 5 |
1890 |
0.3 |
| chr6_32780587_32780883 | 0.65 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
23257 |
0.21 |
| chr3_44001981_44002132 | 0.65 |
Gm37746 |
predicted gene, 37746 |
46335 |
0.18 |
| chr2_79511880_79512425 | 0.65 |
Cerkl |
ceramide kinase-like |
55367 |
0.13 |
| chr11_49185940_49186568 | 0.65 |
Btnl9 |
butyrophilin-like 9 |
783 |
0.5 |
| chr7_83881977_83882131 | 0.64 |
Tlnrd1 |
talin rod domain containing 1 |
2251 |
0.16 |
| chr18_80608643_80609126 | 0.64 |
Nfatc1 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
91 |
0.97 |
| chr6_5005475_5006221 | 0.64 |
Gm26556 |
predicted gene, 26556 |
46865 |
0.12 |
| chr2_168589071_168590755 | 0.64 |
Nfatc2 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
278 |
0.93 |
| chr9_25255721_25255939 | 0.64 |
Septin7 |
septin 7 |
3164 |
0.23 |
| chr10_12019489_12019746 | 0.64 |
Gm48721 |
predicted gene, 48721 |
9846 |
0.2 |
| chr8_40964050_40964403 | 0.64 |
Mtus1 |
mitochondrial tumor suppressor 1 |
35979 |
0.12 |
| chr13_107100937_107101851 | 0.63 |
Gm31452 |
predicted gene, 31452 |
37699 |
0.14 |
| chr15_88758591_88758769 | 0.63 |
Zbed4 |
zinc finger, BED type containing 4 |
6920 |
0.18 |
| chr5_90772565_90772911 | 0.63 |
Pf4 |
platelet factor 4 |
153 |
0.91 |
| chr10_19613570_19613755 | 0.63 |
Ifngr1 |
interferon gamma receptor 1 |
7743 |
0.17 |
| chr11_116108498_116109194 | 0.62 |
Trim47 |
tripartite motif-containing 47 |
727 |
0.48 |
| chr3_8830830_8831235 | 0.62 |
Gm15467 |
predicted gene 15467 |
50167 |
0.11 |
| chr6_143414967_143415230 | 0.62 |
Gm23272 |
predicted gene, 23272 |
50299 |
0.16 |
| chrX_99042650_99043872 | 0.61 |
Stard8 |
START domain containing 8 |
680 |
0.74 |
| chr13_110400077_110400840 | 0.61 |
Plk2 |
polo like kinase 2 |
2661 |
0.34 |
| chr19_11484214_11484365 | 0.61 |
BE692007 |
expressed sequence BE692007 |
246 |
0.82 |
| chr15_55120737_55121109 | 0.61 |
Gm9920 |
predicted gene 9920 |
7246 |
0.16 |
| chr4_139853168_139853461 | 0.61 |
Pax7 |
paired box 7 |
19786 |
0.18 |
| chr7_19011655_19012657 | 0.61 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
8112 |
0.07 |
| chr15_7222960_7224074 | 0.61 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
448 |
0.88 |
| chr18_61785451_61786937 | 0.61 |
Afap1l1 |
actin filament associated protein 1-like 1 |
417 |
0.83 |
| chr8_68556575_68557433 | 0.60 |
Csgalnact1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
27117 |
0.2 |
| chr10_111594457_111595800 | 0.60 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
855 |
0.55 |
| chr6_142980378_142980752 | 0.60 |
A930014E10Rik |
RIKEN cDNA A930014E10 gene |
8813 |
0.14 |
| chr12_21755400_21755551 | 0.60 |
Mrto4-ps2 |
mRNA turnover 4, pseudogene 2 |
21232 |
0.25 |
| chr13_63247351_63247556 | 0.60 |
Gm47586 |
predicted gene, 47586 |
3232 |
0.13 |
| chrX_134275786_134276085 | 0.60 |
Trmt2b |
TRM2 tRNA methyltransferase 2B |
985 |
0.51 |
| chr3_143742898_143743955 | 0.59 |
Gm42705 |
predicted gene 42705 |
49 |
0.97 |
| chr5_62764802_62765190 | 0.59 |
Arap2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
1124 |
0.61 |
| chrX_57337845_57338237 | 0.59 |
Arhgef6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
465 |
0.83 |
| chr2_127728515_127728972 | 0.59 |
Mall |
mal, T cell differentiation protein-like |
1189 |
0.38 |
| chr10_77449173_77450128 | 0.59 |
Gm35920 |
predicted gene, 35920 |
8267 |
0.17 |
| chr7_34745303_34745454 | 0.59 |
Chst8 |
carbohydrate sulfotransferase 8 |
8434 |
0.21 |
| chr17_69121690_69122267 | 0.58 |
Epb41l3 |
erythrocyte membrane protein band 4.1 like 3 |
5022 |
0.32 |
| chr8_85360708_85361016 | 0.58 |
Mylk3 |
myosin light chain kinase 3 |
4462 |
0.15 |
| chr8_34622034_34622185 | 0.58 |
Gm34096 |
predicted gene, 34096 |
13804 |
0.17 |
| chr16_78456546_78456702 | 0.58 |
Gm49603 |
predicted gene, 49603 |
23915 |
0.15 |
| chr16_23993405_23994148 | 0.58 |
Bcl6 |
B cell leukemia/lymphoma 6 |
4924 |
0.18 |
| chr7_96899195_96899433 | 0.58 |
Gm22226 |
predicted gene, 22226 |
20788 |
0.15 |
| chr3_90614243_90615549 | 0.58 |
S100a6 |
S100 calcium binding protein A6 (calcyclin) |
1136 |
0.26 |
| chr2_76808432_76808950 | 0.57 |
Ttn |
titin |
17851 |
0.23 |
| chr5_88928873_88929323 | 0.57 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
5434 |
0.28 |
| chrX_42540237_42540930 | 0.57 |
Sh2d1a |
SH2 domain containing 1A |
13998 |
0.25 |
| chr17_34693537_34693688 | 0.56 |
Gm24389 |
predicted gene, 24389 |
5810 |
0.07 |
| chr15_54413168_54413466 | 0.56 |
Colec10 |
collectin sub-family member 10 |
2543 |
0.32 |
| chr1_168423148_168423974 | 0.56 |
Pbx1 |
pre B cell leukemia homeobox 1 |
7943 |
0.26 |
| chr2_176121352_176121707 | 0.56 |
Gm16357 |
predicted gene 16357 |
3663 |
0.21 |
| chr14_54959915_54960463 | 0.55 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
2282 |
0.1 |
| chr1_189340106_189340450 | 0.55 |
Kcnk2 |
potassium channel, subfamily K, member 2 |
3077 |
0.24 |
| chr4_15883184_15883580 | 0.55 |
Calb1 |
calbindin 1 |
2075 |
0.27 |
| chr4_106489848_106489999 | 0.55 |
Bsnd |
barttin CLCNK type accessory beta subunit |
2360 |
0.2 |
| chr15_5927691_5927842 | 0.55 |
Gm49095 |
predicted gene, 49095 |
23678 |
0.2 |
| chr6_74588312_74588463 | 0.55 |
Gm22254 |
predicted gene, 22254 |
83223 |
0.11 |
| chr4_143238489_143238893 | 0.55 |
Eef2-ps2 |
eukaryotic translation elongation factor 2, pseudogene 2 |
4677 |
0.17 |
| chr12_36137152_36137570 | 0.54 |
Gm48369 |
predicted gene, 48369 |
10561 |
0.12 |
| chr4_16168304_16169243 | 0.54 |
A530072M11Rik |
RIKEN cDNA gene A530072M11 |
4663 |
0.18 |
| chr2_177032666_177033048 | 0.54 |
Gm14402 |
predicted gene 14402 |
202 |
0.91 |
| chr15_7145070_7145221 | 0.54 |
Lifr |
LIF receptor alpha |
4596 |
0.31 |
| chr5_24365290_24365969 | 0.54 |
Nos3 |
nitric oxide synthase 3, endothelial cell |
819 |
0.41 |
| chr6_59022486_59023190 | 0.53 |
Fam13a |
family with sequence similarity 13, member A |
1502 |
0.36 |
| chr10_4611597_4612860 | 0.53 |
Esr1 |
estrogen receptor 1 (alpha) |
207 |
0.95 |
| chr12_40381138_40381829 | 0.53 |
Zfp277 |
zinc finger protein 277 |
18599 |
0.22 |
| chr1_43182605_43182756 | 0.53 |
Gm8210 |
predicted pseudogene 8210 |
6476 |
0.19 |
| chr6_86480161_86480975 | 0.53 |
A430078I02Rik |
RIKEN cDNA A430078I02 gene |
202 |
0.86 |
| chr10_68541127_68542141 | 0.53 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
262 |
0.94 |
| chr11_89066018_89066308 | 0.52 |
Dgkeos |
diacylglycerol kinase, epsilon, opposite strand |
75 |
0.91 |
| chr3_101598645_101599448 | 0.52 |
Gm42941 |
predicted gene 42941 |
1916 |
0.31 |
| chr8_11331313_11331580 | 0.52 |
Col4a1 |
collagen, type IV, alpha 1 |
18620 |
0.15 |
| chr10_5287995_5289689 | 0.52 |
Gm23573 |
predicted gene, 23573 |
68331 |
0.12 |
| chr6_91960417_91960897 | 0.52 |
4930590J08Rik |
RIKEN cDNA 4930590J08 gene |
16393 |
0.13 |
| chr3_152191751_152192635 | 0.52 |
Dnajb4 |
DnaJ heat shock protein family (Hsp40) member B4 |
1652 |
0.26 |
| chr4_141304021_141304172 | 0.52 |
Epha2 |
Eph receptor A2 |
1689 |
0.23 |
| chr17_45620159_45620310 | 0.52 |
Gm25008 |
predicted gene, 25008 |
445 |
0.69 |
| chr15_89425017_89425997 | 0.51 |
Cpt1b |
carnitine palmitoyltransferase 1b, muscle |
288 |
0.74 |
| chr3_40509511_40509727 | 0.51 |
1700017G19Rik |
RIKEN cDNA 1700017G19 gene |
155 |
0.96 |
| chr8_77523888_77524583 | 0.51 |
0610038B21Rik |
RIKEN cDNA 0610038B21 gene |
6120 |
0.15 |
| chr15_91028535_91029324 | 0.51 |
Kif21a |
kinesin family member 21A |
20889 |
0.18 |
| chr2_176831033_176831392 | 0.51 |
Gm14408 |
predicted gene 14408 |
72 |
0.97 |
| chr3_41173045_41173450 | 0.51 |
Gm40038 |
predicted gene, 40038 |
584 |
0.8 |
| chr7_132656285_132656853 | 0.51 |
Gm15582 |
predicted gene 15582 |
3315 |
0.23 |
| chr4_63151303_63151507 | 0.51 |
Ambp |
alpha 1 microglobulin/bikunin precursor |
2768 |
0.24 |
| chr15_40354440_40354591 | 0.51 |
Gm41311 |
predicted gene, 41311 |
11255 |
0.28 |
| chr1_194940483_194941153 | 0.51 |
Cd34 |
CD34 antigen |
1823 |
0.26 |
| chr16_31695067_31695951 | 0.51 |
Dlg1 |
discs large MAGUK scaffold protein 1 |
11243 |
0.15 |
| chr14_119260402_119261033 | 0.51 |
Gm6087 |
predicted gene 6087 |
17707 |
0.26 |
| chr7_135604964_135605508 | 0.50 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
592 |
0.74 |
| chr9_69092316_69092646 | 0.50 |
Rora |
RAR-related orphan receptor alpha |
103060 |
0.08 |
| chr13_46503428_46503590 | 0.50 |
Cap2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
1343 |
0.34 |
| chr12_19201995_19202146 | 0.50 |
Gm48528 |
predicted gene, 48528 |
16012 |
0.14 |
| chr17_26503687_26504320 | 0.50 |
Gm34455 |
predicted gene, 34455 |
803 |
0.47 |
| chr13_59970938_59972210 | 0.50 |
Gm48390 |
predicted gene, 48390 |
52136 |
0.09 |
| chr3_38015645_38016389 | 0.50 |
Gm22899 |
predicted gene, 22899 |
20782 |
0.16 |
| chr13_38602280_38602560 | 0.50 |
Gm47400 |
predicted gene, 47400 |
32551 |
0.1 |
| chr5_124960229_124960932 | 0.50 |
Rflna |
refilin A |
42641 |
0.13 |
| chr5_77579867_77580744 | 0.50 |
Pea15b |
phosphoprotein enriched in astrocytes 15B |
68557 |
0.09 |
| chr9_70567831_70568043 | 0.49 |
Sltm |
SAFB-like, transcription modulator |
7471 |
0.16 |
| chr3_93568427_93569206 | 0.48 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
8089 |
0.13 |
| chr6_88189310_88190735 | 0.48 |
Gm38708 |
predicted gene, 38708 |
659 |
0.6 |
| chr14_93306296_93306447 | 0.48 |
Gm48964 |
predicted gene, 48964 |
4766 |
0.32 |
| chr10_89453189_89453411 | 0.48 |
Gas2l3 |
growth arrest-specific 2 like 3 |
9333 |
0.24 |
| chr10_95725993_95726144 | 0.48 |
1110019B22Rik |
RIKEN cDNA 1110019B22 gene |
3568 |
0.14 |
| chr8_11260160_11260311 | 0.48 |
Col4a1 |
collagen, type IV, alpha 1 |
14630 |
0.16 |
| chr5_81509100_81509275 | 0.47 |
Gm43084 |
predicted gene 43084 |
26539 |
0.26 |
| chr14_101886945_101887764 | 0.47 |
Lmo7 |
LIM domain only 7 |
3235 |
0.35 |
| chr9_118488682_118489383 | 0.47 |
Gm33460 |
predicted gene, 33460 |
67 |
0.96 |
| chr11_78984345_78985263 | 0.47 |
Lgals9 |
lectin, galactose binding, soluble 9 |
27 |
0.98 |
| chr2_68435653_68436705 | 0.47 |
Stk39 |
serine/threonine kinase 39 |
35761 |
0.16 |
| chr2_131129425_131129576 | 0.47 |
Hspa12b |
heat shock protein 12B |
2006 |
0.18 |
| chr9_32341364_32342112 | 0.47 |
Kcnj5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
2499 |
0.25 |
| chr6_23462582_23462958 | 0.47 |
Gm27959 |
predicted gene, 27959 |
56976 |
0.14 |
| chr19_53325039_53326410 | 0.46 |
Gm30541 |
predicted gene, 30541 |
1 |
0.97 |
| chr17_86364752_86364903 | 0.46 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
77649 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.3 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.3 | 0.8 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 0.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.2 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 0.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.8 | GO:1900212 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.2 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 0.5 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.2 | 0.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.1 | 1.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.3 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.1 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.2 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.3 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.2 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.2 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.1 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.3 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.3 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.3 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.5 | GO:1901739 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.3 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.5 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.0 | GO:1900126 | regulation of hyaluronan biosynthetic process(GO:1900125) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 0.0 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.3 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 1.1 | GO:0050818 | regulation of coagulation(GO:0050818) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.0 | GO:0060558 | regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.0 | 0.0 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.1 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.0 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.0 | GO:1902475 | L-glutamate transmembrane transport(GO:0089711) L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.0 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.0 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 2.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.6 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.2 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0043731 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.0 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME SEMAPHORIN INTERACTIONS | Genes involved in Semaphorin interactions |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |