Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
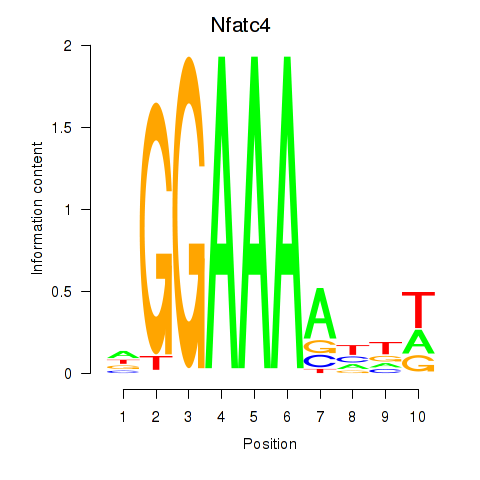
Results for Nfatc4
Z-value: 0.19
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.5 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
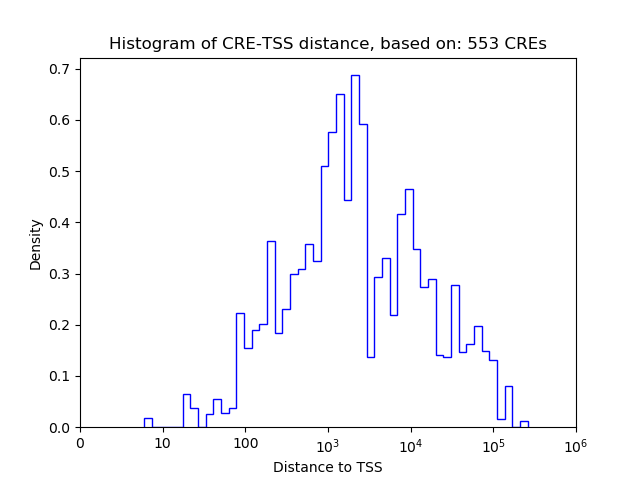
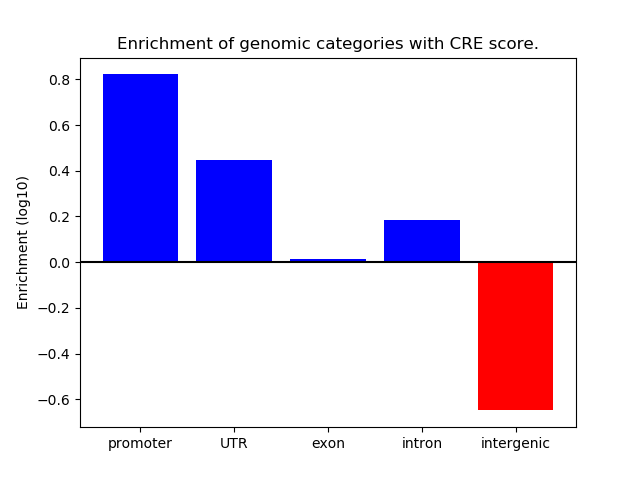
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_55828255_55828946 | Nfatc4 | 869 | 0.391501 | 0.32 | 1.3e-02 | Click! |
| chr14_55829268_55829478 | Nfatc4 | 96 | 0.931307 | 0.30 | 1.9e-02 | Click! |
| chr14_55824784_55826081 | Nfatc4 | 369 | 0.734082 | 0.26 | 4.1e-02 | Click! |
| chr14_55822882_55823033 | Nfatc4 | 187 | 0.880957 | 0.24 | 6.4e-02 | Click! |
| chr14_55831685_55831836 | Nfatc4 | 1491 | 0.222718 | 0.23 | 8.0e-02 | Click! |
Activity of the Nfatc4 motif across conditions
Conditions sorted by the z-value of the Nfatc4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
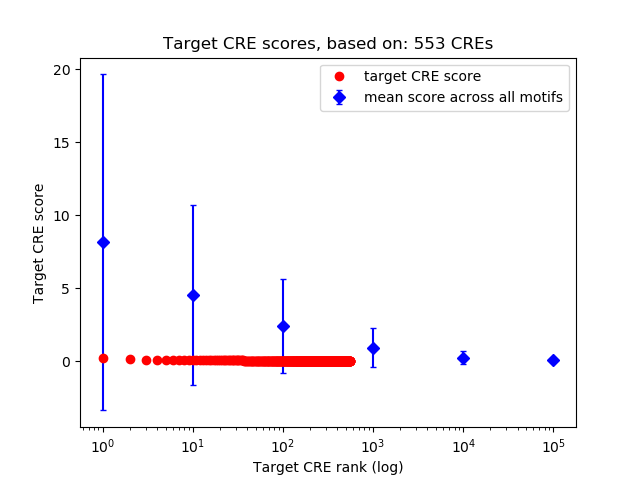
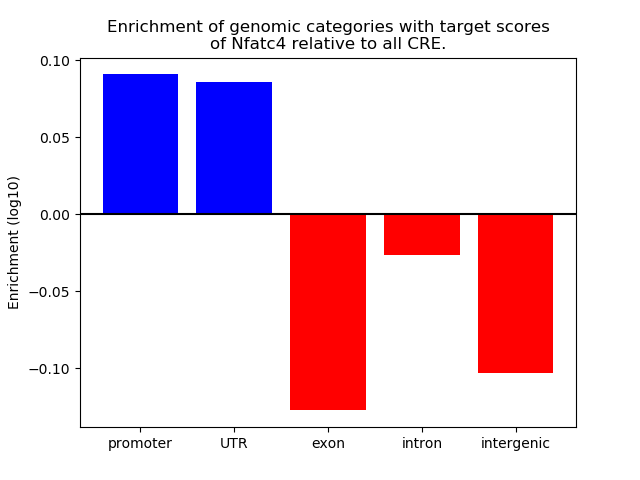
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_74689028_74690409 | 0.21 |
Hoxd10 |
homeobox D10 |
2206 |
0.09 |
| chr4_156057650_156058246 | 0.14 |
Ttll10 |
tubulin tyrosine ligase-like family, member 10 |
1396 |
0.16 |
| chr8_33926179_33926910 | 0.11 |
Rbpms |
RNA binding protein gene with multiple splicing |
2732 |
0.25 |
| chr9_75773918_75774501 | 0.10 |
Bmp5 |
bone morphogenetic protein 5 |
1155 |
0.53 |
| chr7_134228173_134228324 | 0.10 |
A130023I24Rik |
RIKEN cDNA A130023I24 gene |
1072 |
0.52 |
| chr6_52174920_52176658 | 0.10 |
Hoxaas3 |
Hoxa cluster antisense RNA 3 |
554 |
0.41 |
| chr5_90519467_90519771 | 0.09 |
Afm |
afamin |
670 |
0.59 |
| chr7_142661369_142661520 | 0.09 |
Igf2 |
insulin-like growth factor 2 |
139 |
0.91 |
| chr11_96340356_96341209 | 0.08 |
Hoxb3 |
homeobox B3 |
163 |
0.85 |
| chr1_78816815_78817194 | 0.08 |
Kcne4 |
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
77 |
0.97 |
| chr6_51401722_51401977 | 0.08 |
0610033M10Rik |
RIKEN cDNA 0610033M10 gene |
184 |
0.94 |
| chr10_105794148_105794388 | 0.08 |
n-R5s80 |
nuclear encoded rRNA 5S 80 |
9929 |
0.18 |
| chr6_48707880_48708301 | 0.07 |
Gimap6 |
GTPase, IMAP family member 6 |
106 |
0.91 |
| chr8_87570471_87571218 | 0.07 |
4933402J07Rik |
RIKEN cDNA 4933402J07 gene |
6991 |
0.21 |
| chr9_45042261_45043690 | 0.07 |
Mpzl2 |
myelin protein zero-like 2 |
279 |
0.8 |
| chr1_184731287_184732512 | 0.07 |
Hlx |
H2.0-like homeobox |
301 |
0.86 |
| chr3_137343040_137343246 | 0.07 |
Emcn |
endomucin |
1969 |
0.42 |
| chr2_169996778_169996974 | 0.07 |
AY702102 |
cDNA sequence AY702102 |
33689 |
0.22 |
| chr4_49545411_49545633 | 0.07 |
Aldob |
aldolase B, fructose-bisphosphate |
4024 |
0.16 |
| chr7_100924536_100925334 | 0.07 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
7172 |
0.15 |
| chr19_5558301_5558859 | 0.06 |
Ovol1 |
ovo like zinc finger 1 |
823 |
0.33 |
| chr7_75457069_75457220 | 0.06 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
1276 |
0.42 |
| chr9_123214842_123215032 | 0.06 |
Cdcp1 |
CUB domain containing protein 1 |
1062 |
0.46 |
| chr13_98626689_98627173 | 0.06 |
Gm48050 |
predicted gene, 48050 |
9269 |
0.13 |
| chr2_35691764_35692937 | 0.06 |
Dab2ip |
disabled 2 interacting protein |
17 |
0.98 |
| chr9_42884724_42884990 | 0.06 |
Gm27509 |
predicted gene, 27509 |
22728 |
0.19 |
| chr16_23102639_23103369 | 0.06 |
Eif4a2 |
eukaryotic translation initiation factor 4A2 |
4440 |
0.07 |
| chr10_10546265_10546583 | 0.06 |
Rab32 |
RAB32, member RAS oncogene family |
11757 |
0.2 |
| chr12_71342419_71342699 | 0.06 |
Gm33016 |
predicted gene, 33016 |
4931 |
0.16 |
| chr5_134342577_134343060 | 0.06 |
Mir3965 |
microRNA 3965 |
16364 |
0.12 |
| chr8_36829881_36830312 | 0.06 |
Dlc1 |
deleted in liver cancer 1 |
97042 |
0.08 |
| chr18_69665931_69667348 | 0.06 |
Tcf4 |
transcription factor 4 |
9972 |
0.29 |
| chr1_45924247_45924747 | 0.06 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
765 |
0.56 |
| chr13_21720851_21721002 | 0.05 |
Gm44357 |
predicted gene, 44357 |
507 |
0.36 |
| chr8_107198114_107198786 | 0.05 |
Cyb5b |
cytochrome b5 type B |
32535 |
0.13 |
| chr13_89540278_89541317 | 0.05 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
1001 |
0.64 |
| chr1_78817332_78818790 | 0.05 |
Kcne4 |
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
1134 |
0.48 |
| chr15_72971199_72971927 | 0.05 |
Gm3150 |
predicted gene 3150 |
58206 |
0.13 |
| chr5_119662786_119663760 | 0.05 |
Tbx3 |
T-box 3 |
7396 |
0.14 |
| chr13_21820445_21820658 | 0.05 |
Hist1h4m |
histone cluster 1, H4m |
8805 |
0.05 |
| chr2_84423814_84425562 | 0.05 |
Calcrl |
calcitonin receptor-like |
578 |
0.75 |
| chr8_92761797_92762731 | 0.05 |
Gapdh-ps16 |
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 16 |
45289 |
0.12 |
| chr4_135727528_135728972 | 0.05 |
Il22ra1 |
interleukin 22 receptor, alpha 1 |
78 |
0.96 |
| chr6_117919030_117919254 | 0.05 |
Gm43863 |
predicted gene, 43863 |
1420 |
0.24 |
| chr10_22822761_22823454 | 0.05 |
Tcf21 |
transcription factor 21 |
2933 |
0.22 |
| chr5_66149768_66149919 | 0.05 |
Rbm47 |
RNA binding motif protein 47 |
1113 |
0.3 |
| chr17_38303471_38303622 | 0.05 |
Olfr137 |
olfactory receptor 137 |
1914 |
0.25 |
| chr6_17309182_17310034 | 0.05 |
Cav1 |
caveolin 1, caveolae protein |
1885 |
0.33 |
| chr8_84654998_84655201 | 0.05 |
Ier2 |
immediate early response 2 |
7755 |
0.1 |
| chr12_104584053_104584862 | 0.05 |
Gm49732 |
predicted gene, 49732 |
64489 |
0.1 |
| chr4_33245981_33246490 | 0.05 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
1340 |
0.4 |
| chr13_104122133_104122507 | 0.05 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
12491 |
0.16 |
| chr6_30544847_30545729 | 0.05 |
Cpa2 |
carboxypeptidase A2, pancreatic |
803 |
0.52 |
| chr9_117249167_117249456 | 0.05 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
2286 |
0.33 |
| chr13_38959403_38959696 | 0.05 |
Slc35b3 |
solute carrier family 35, member B3 |
931 |
0.42 |
| chr6_138143538_138143877 | 0.05 |
Mgst1 |
microsomal glutathione S-transferase 1 |
853 |
0.73 |
| chr6_5255342_5256653 | 0.05 |
Pon3 |
paraoxonase 3 |
217 |
0.93 |
| chr11_98904753_98905026 | 0.05 |
Cdc6 |
cell division cycle 6 |
2912 |
0.15 |
| chr2_34774288_34775191 | 0.05 |
Hspa5 |
heat shock protein 5 |
108 |
0.95 |
| chr16_50752363_50752789 | 0.04 |
Dubr |
Dppa2 upstream binding RNA |
19803 |
0.17 |
| chr4_115001106_115001269 | 0.04 |
Stil |
Scl/Tal1 interrupting locus |
1005 |
0.49 |
| chr3_53055538_53055689 | 0.04 |
Gm42901 |
predicted gene 42901 |
13858 |
0.11 |
| chr2_137109115_137109266 | 0.04 |
Jag1 |
jagged 1 |
7454 |
0.29 |
| chr17_5753255_5753544 | 0.04 |
3300005D01Rik |
RIKEN cDNA 3300005D01 gene |
45258 |
0.13 |
| chr13_43123788_43124308 | 0.04 |
Phactr1 |
phosphatase and actin regulator 1 |
1176 |
0.42 |
| chr19_55260209_55260360 | 0.04 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
6915 |
0.18 |
| chrX_106483380_106483727 | 0.04 |
Fndc3c1 |
fibronectin type III domain containing 3C1 |
1344 |
0.43 |
| chr15_98606352_98606619 | 0.04 |
Adcy6 |
adenylate cyclase 6 |
1148 |
0.29 |
| chr16_44348732_44349059 | 0.04 |
Spice1 |
spindle and centriole associated protein 1 |
849 |
0.63 |
| chr13_115054607_115055088 | 0.04 |
Gm10734 |
predicted gene 10734 |
34654 |
0.12 |
| chr5_90773168_90773319 | 0.04 |
Pf4 |
platelet factor 4 |
352 |
0.76 |
| chr16_31575640_31575823 | 0.04 |
Gm34256 |
predicted gene, 34256 |
17489 |
0.16 |
| chr2_103283203_103283868 | 0.04 |
Ehf |
ets homologous factor |
225 |
0.93 |
| chr2_156324321_156324820 | 0.04 |
Gm22759 |
predicted gene, 22759 |
2018 |
0.15 |
| chr9_89497806_89497957 | 0.04 |
Gm47403 |
predicted gene, 47403 |
62503 |
0.11 |
| chr10_44002146_44002297 | 0.04 |
Gm10812 |
predicted gene 10812 |
1933 |
0.28 |
| chr13_40136054_40136205 | 0.04 |
Ofcc1 |
orofacial cleft 1 candidate 1 |
151882 |
0.04 |
| chr11_40734727_40734972 | 0.04 |
Nudcd2 |
NudC domain containing 2 |
644 |
0.6 |
| chr7_127769289_127770591 | 0.04 |
Orai3 |
ORAI calcium release-activated calcium modulator 3 |
125 |
0.9 |
| chr1_39574806_39574957 | 0.04 |
Gm23722 |
predicted gene, 23722 |
1006 |
0.38 |
| chr19_5257020_5257171 | 0.04 |
Gm50189 |
predicted gene, 50189 |
1655 |
0.19 |
| chr11_98412611_98413343 | 0.04 |
Erbb2 |
erb-b2 receptor tyrosine kinase 2 |
484 |
0.6 |
| chr5_140890731_140891173 | 0.04 |
Card11 |
caspase recruitment domain family, member 11 |
8747 |
0.24 |
| chr14_22049015_22049482 | 0.04 |
Gm7480 |
predicted gene 7480 |
13145 |
0.16 |
| chr2_35060883_35061388 | 0.04 |
Hc |
hemolytic complement |
303 |
0.88 |
| chr11_117611027_117611290 | 0.04 |
2900041M22Rik |
RIKEN cDNA 2900041M22 gene |
89 |
0.97 |
| chr1_39636608_39636940 | 0.04 |
D930019O06Rik |
RIKEN cDNA D930019O06 |
13402 |
0.13 |
| chr5_59968294_59968445 | 0.04 |
Gm43041 |
predicted gene 43041 |
12229 |
0.17 |
| chr5_20941736_20941887 | 0.04 |
Rsbn1l |
round spermatid basic protein 1-like |
3693 |
0.2 |
| chr15_80674232_80674436 | 0.04 |
Fam83f |
family with sequence similarity 83, member F |
2487 |
0.2 |
| chr7_96533226_96533480 | 0.04 |
Tenm4 |
teneurin transmembrane protein 4 |
10955 |
0.26 |
| chr3_100479788_100480010 | 0.04 |
Tent5c |
terminal nucleotidyltransferase 5C |
9295 |
0.14 |
| chr1_121096624_121097922 | 0.04 |
Celrr |
cerebellum expressed regulatory RNA |
9744 |
0.27 |
| chr8_128688078_128688272 | 0.04 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2305 |
0.29 |
| chr5_147299313_147300749 | 0.04 |
Cdx2 |
caudal type homeobox 2 |
7239 |
0.1 |
| chr8_45627436_45629381 | 0.04 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
207 |
0.94 |
| chr8_109360134_109360931 | 0.04 |
Gm1943 |
predicted gene 1943 |
19668 |
0.22 |
| chr6_138140044_138141370 | 0.04 |
Mgst1 |
microsomal glutathione S-transferase 1 |
108 |
0.98 |
| chr18_61045132_61047010 | 0.04 |
Pdgfrb |
platelet derived growth factor receptor, beta polypeptide |
871 |
0.5 |
| chr5_26075770_26075952 | 0.04 |
Gm6089 |
predicted gene 6089 |
5324 |
0.12 |
| chr2_139634376_139634527 | 0.04 |
Gm14066 |
predicted gene 14066 |
43263 |
0.14 |
| chr10_93454496_93455097 | 0.04 |
Lta4h |
leukotriene A4 hydrolase |
1345 |
0.38 |
| chr10_87524981_87525132 | 0.04 |
Pah |
phenylalanine hydroxylase |
3029 |
0.26 |
| chr6_83032569_83034015 | 0.04 |
Dok1 |
docking protein 1 |
176 |
0.8 |
| chr14_65951762_65951913 | 0.04 |
Scara3 |
scavenger receptor class A, member 3 |
2098 |
0.27 |
| chr2_148023997_148024148 | 0.04 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
14198 |
0.17 |
| chr10_87007185_87007640 | 0.04 |
Stab2 |
stabilin 2 |
600 |
0.7 |
| chr13_21452056_21452233 | 0.04 |
Zscan26 |
zinc finger and SCAN domain containing 26 |
1516 |
0.16 |
| chr3_59129690_59130148 | 0.04 |
P2ry14 |
purinergic receptor P2Y, G-protein coupled, 14 |
703 |
0.66 |
| chr13_23582405_23582731 | 0.03 |
H4c4 |
H4 clustered histone 4 |
970 |
0.2 |
| chr12_78205797_78206679 | 0.03 |
Gm6657 |
predicted gene 6657 |
5272 |
0.17 |
| chr3_152212912_152213690 | 0.03 |
Fubp1 |
far upstream element (FUSE) binding protein 1 |
2780 |
0.17 |
| chr7_143685249_143686318 | 0.03 |
Tnfrsf23 |
tumor necrosis factor receptor superfamily, member 23 |
88 |
0.95 |
| chr8_11779325_11780029 | 0.03 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
2850 |
0.19 |
| chr17_43395854_43396005 | 0.03 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
6441 |
0.25 |
| chr5_32611887_32612080 | 0.03 |
Gm10461 |
predicted gene 10461 |
178 |
0.83 |
| chr1_189730413_189731175 | 0.03 |
Ptpn14 |
protein tyrosine phosphatase, non-receptor type 14 |
2519 |
0.29 |
| chr10_93347628_93348351 | 0.03 |
Gm17745 |
predicted gene, 17745 |
12596 |
0.16 |
| chr6_117130561_117131249 | 0.03 |
Gm43930 |
predicted gene, 43930 |
13420 |
0.19 |
| chr8_53637994_53639312 | 0.03 |
Neil3 |
nei like 3 (E. coli) |
25 |
0.99 |
| chrX_12286250_12287038 | 0.03 |
Gm14635 |
predicted gene 14635 |
58578 |
0.14 |
| chr6_125426842_125427196 | 0.03 |
Cd9 |
CD9 antigen |
36005 |
0.1 |
| chr1_134992164_134992455 | 0.03 |
Ube2t |
ubiquitin-conjugating enzyme E2T |
20353 |
0.15 |
| chr11_49246331_49246550 | 0.03 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
1037 |
0.4 |
| chr12_73287843_73287994 | 0.03 |
Slc38a6 |
solute carrier family 38, member 6 |
858 |
0.48 |
| chr12_53408081_53408232 | 0.03 |
1700030L22Rik |
RIKEN cDNA 1700030L22 gene |
98310 |
0.09 |
| chr2_74722101_74723871 | 0.03 |
Hoxd4 |
homeobox D4 |
1008 |
0.22 |
| chrY_1246020_1246180 | 0.03 |
Uty |
ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome |
341 |
0.86 |
| chr2_177839442_177839866 | 0.03 |
Gm14325 |
predicted gene 14325 |
664 |
0.67 |
| chr4_84672964_84673574 | 0.03 |
Bnc2 |
basonuclin 2 |
1727 |
0.42 |
| chr18_63689730_63689974 | 0.03 |
Txnl1 |
thioredoxin-like 1 |
2470 |
0.26 |
| chr10_21995510_21995970 | 0.03 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
830 |
0.57 |
| chr6_114875505_114875725 | 0.03 |
Vgll4 |
vestigial like family member 4 |
508 |
0.81 |
| chr6_119416453_119417745 | 0.03 |
Adipor2 |
adiponectin receptor 2 |
376 |
0.88 |
| chrX_53058333_53059722 | 0.03 |
C430049B03Rik |
RIKEN cDNA C430049B03 gene |
1837 |
0.16 |
| chr2_84713270_84713499 | 0.03 |
Zdhhc5 |
zinc finger, DHHC domain containing 5 |
1302 |
0.22 |
| chr4_46401911_46402304 | 0.03 |
Hemgn |
hemogen |
2129 |
0.22 |
| chr12_68675940_68676091 | 0.03 |
n-R5s61 |
nuclear encoded rRNA 5S 61 |
75045 |
0.09 |
| chr2_138264683_138264834 | 0.03 |
Btbd3 |
BTB (POZ) domain containing 3 |
8088 |
0.33 |
| chr5_130022581_130023074 | 0.03 |
Asl |
argininosuccinate lyase |
1473 |
0.28 |
| chr6_120195255_120195624 | 0.03 |
Ninj2 |
ninjurin 2 |
1616 |
0.41 |
| chr17_80825543_80826587 | 0.03 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
70728 |
0.1 |
| chr8_68279205_68280180 | 0.03 |
Sh2d4a |
SH2 domain containing 4A |
2615 |
0.27 |
| chr2_180512043_180512194 | 0.03 |
Ntsr1 |
neurotensin receptor 1 |
11891 |
0.16 |
| chr2_29546441_29547087 | 0.03 |
Med27 |
mediator complex subunit 27 |
41893 |
0.12 |
| chr10_60563030_60563532 | 0.03 |
Cdh23 |
cadherin 23 (otocadherin) |
89752 |
0.08 |
| chr14_78012288_78012541 | 0.03 |
Gm48954 |
predicted gene, 48954 |
7314 |
0.21 |
| chr1_150898005_150898378 | 0.03 |
Hmcn1 |
hemicentin 1 |
94860 |
0.07 |
| chr8_123102029_123102227 | 0.03 |
Rpl13 |
ribosomal protein L13 |
222 |
0.79 |
| chr16_72719540_72719691 | 0.03 |
Robo1 |
roundabout guidance receptor 1 |
56411 |
0.18 |
| chr4_123233207_123234595 | 0.03 |
Heyl |
hairy/enhancer-of-split related with YRPW motif-like |
48 |
0.96 |
| chr10_99267189_99269284 | 0.03 |
Gm48089 |
predicted gene, 48089 |
340 |
0.78 |
| chr4_3937146_3938329 | 0.03 |
Plag1 |
pleiomorphic adenoma gene 1 |
645 |
0.54 |
| chr2_128377708_128377954 | 0.03 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
25134 |
0.18 |
| chr7_27596549_27597871 | 0.03 |
Akt2 |
thymoma viral proto-oncogene 2 |
380 |
0.77 |
| chr17_71236074_71236240 | 0.03 |
Lpin2 |
lipin 2 |
2505 |
0.25 |
| chr6_75117264_75117415 | 0.03 |
Gm8992 |
predicted gene 8992 |
70829 |
0.12 |
| chr6_108235668_108236315 | 0.03 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
16967 |
0.23 |
| chr9_65574695_65575665 | 0.03 |
Plekho2 |
pleckstrin homology domain containing, family O member 2 |
1920 |
0.26 |
| chr15_39078038_39078699 | 0.03 |
Cthrc1 |
collagen triple helix repeat containing 1 |
1287 |
0.33 |
| chr11_102609159_102609359 | 0.03 |
Fzd2 |
frizzled class receptor 2 |
4863 |
0.11 |
| chr12_69757617_69759559 | 0.03 |
Mir681 |
microRNA 681 |
5356 |
0.14 |
| chr17_19455750_19455901 | 0.03 |
Vmn2r-ps121 |
vomeronasal 2, receptor, pseudogene 121 |
2500 |
0.3 |
| chr4_100863972_100864540 | 0.03 |
Cachd1 |
cache domain containing 1 |
87581 |
0.09 |
| chr10_98624940_98625347 | 0.03 |
Gm5427 |
predicted gene 5427 |
74567 |
0.11 |
| chr3_90220236_90221599 | 0.03 |
Rab13 |
RAB13, member RAS oncogene family |
130 |
0.91 |
| chr4_134766990_134768275 | 0.03 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
392 |
0.86 |
| chr2_79256898_79257323 | 0.03 |
Itga4 |
integrin alpha 4 |
1163 |
0.57 |
| chr1_168754197_168754399 | 0.03 |
1700063I16Rik |
RIKEN cDNA 1700063I16 gene |
78416 |
0.12 |
| chr11_76997745_76998893 | 0.03 |
Slc6a4 |
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
284 |
0.87 |
| chr15_77142814_77142965 | 0.03 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
10109 |
0.14 |
| chr13_78629090_78629413 | 0.03 |
Gm48402 |
predicted gene, 48402 |
8766 |
0.3 |
| chr12_100998948_100999342 | 0.03 |
Gm36756 |
predicted gene, 36756 |
6173 |
0.13 |
| chr3_109025772_109026235 | 0.03 |
Fam102b |
family with sequence similarity 102, member B |
1270 |
0.37 |
| chr7_29799575_29799936 | 0.03 |
Gm6579 |
predicted gene 6579 |
41 |
0.95 |
| chr13_63560468_63561467 | 0.03 |
Ptch1 |
patched 1 |
2848 |
0.2 |
| chrX_169685204_169686251 | 0.03 |
Mid1 |
midline 1 |
528 |
0.82 |
| chr13_88872554_88872705 | 0.03 |
Edil3 |
EGF-like repeats and discoidin I-like domains 3 |
50987 |
0.19 |
| chr7_116267794_116268386 | 0.03 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
3662 |
0.2 |
| chr16_95618168_95618511 | 0.03 |
Erg |
ETS transcription factor |
31746 |
0.2 |
| chr4_126490666_126491063 | 0.03 |
Gm12928 |
predicted gene 12928 |
13565 |
0.1 |
| chr7_141442950_141444299 | 0.03 |
Pidd1 |
p53 induced death domain protein 1 |
268 |
0.74 |
| chr7_73606673_73607479 | 0.03 |
Gm44734 |
predicted gene 44734 |
1872 |
0.21 |
| chr11_100702726_100703447 | 0.03 |
Dhx58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
261 |
0.82 |
| chr6_52273154_52274564 | 0.03 |
Hottip |
Hoxa distal transcript antisense RNA |
11075 |
0.06 |
| chr14_109083684_109083835 | 0.03 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
169601 |
0.04 |
| chr14_27337217_27337907 | 0.03 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
1496 |
0.45 |
| chr14_36918744_36919600 | 0.03 |
Ccser2 |
coiled-coil serine rich 2 |
161 |
0.97 |
| chr15_56694253_56695303 | 0.03 |
Has2 |
hyaluronan synthase 2 |
239 |
0.85 |
| chrX_9467520_9467766 | 0.03 |
Cybb |
cytochrome b-245, beta polypeptide |
1626 |
0.4 |
| chr1_164062190_164062341 | 0.03 |
Sell |
selectin, lymphocyte |
76 |
0.96 |
| chr6_56704794_56705009 | 0.03 |
Lsm5 |
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
191 |
0.94 |
| chr7_84688773_84688924 | 0.03 |
2610206C17Rik |
RIKEN cDNA 2610206C17 gene |
792 |
0.47 |
| chr11_102603430_102604064 | 0.03 |
Fzd2 |
frizzled class receptor 2 |
649 |
0.54 |
| chr13_72625092_72626461 | 0.03 |
Gm20554 |
predicted gene, 20554 |
2788 |
0.27 |
| chr4_116166621_116166961 | 0.03 |
Tspan1 |
tetraspanin 1 |
809 |
0.42 |
| chr2_127729042_127730365 | 0.03 |
Mall |
mal, T cell differentiation protein-like |
229 |
0.9 |
| chr8_45261536_45262005 | 0.03 |
F11 |
coagulation factor XI |
261 |
0.91 |
| chr8_79030497_79030682 | 0.03 |
Zfp827 |
zinc finger protein 827 |
2002 |
0.34 |
| chr10_37139080_37141738 | 0.02 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
495 |
0.74 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |