Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
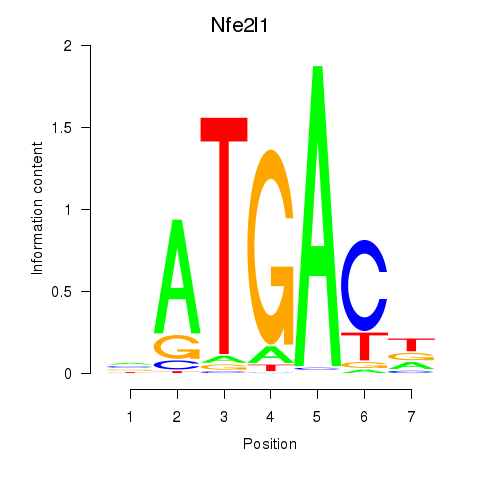
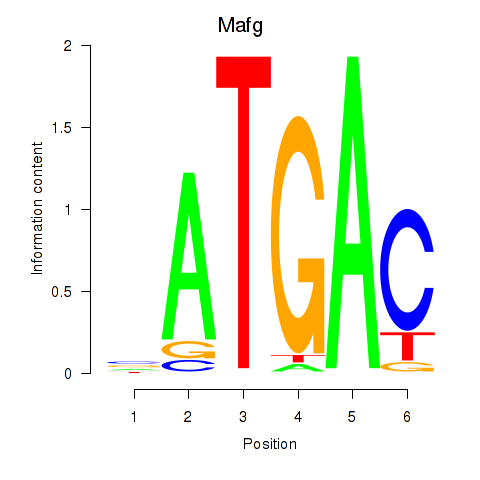
Results for Nfe2l1_Mafg
Z-value: 0.40
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.11 | nuclear factor, erythroid derived 2,-like 1 |
|
Mafg
|
ENSMUSG00000051510.7 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
|
Mafg
|
ENSMUSG00000053906.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_120635927_120636078 | Mafg | 2402 | 0.087462 | -0.40 | 1.4e-03 | Click! |
| chr11_120637299_120637552 | Mafg | 3825 | 0.065678 | -0.19 | 1.5e-01 | Click! |
| chr11_120633726_120633919 | Mafg | 222 | 0.773977 | 0.16 | 2.2e-01 | Click! |
| chr11_120632083_120632234 | Mafg | 1361 | 0.154242 | 0.15 | 2.4e-01 | Click! |
| chr11_120628347_120628498 | Mafg | 1726 | 0.114680 | 0.12 | 3.5e-01 | Click! |
| chr11_96837393_96837544 | Nfe2l1 | 7500 | 0.097092 | -0.45 | 3.4e-04 | Click! |
| chr11_96818178_96818461 | Nfe2l1 | 4406 | 0.116272 | -0.39 | 2.3e-03 | Click! |
| chr11_96830243_96830655 | Nfe2l1 | 481 | 0.660634 | 0.29 | 2.4e-02 | Click! |
| chr11_96828944_96829908 | Nfe2l1 | 75 | 0.943784 | 0.22 | 8.5e-02 | Click! |
| chr11_96823513_96824552 | Nfe2l1 | 19 | 0.954431 | -0.21 | 1.0e-01 | Click! |
Activity of the Nfe2l1_Mafg motif across conditions
Conditions sorted by the z-value of the Nfe2l1_Mafg motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
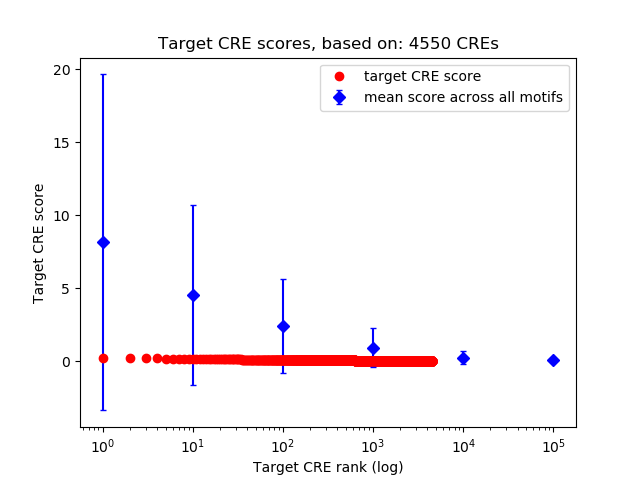
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_75577269_75578512 | 0.25 |
Kit |
KIT proto-oncogene receptor tyrosine kinase |
2935 |
0.29 |
| chr13_109928298_109928867 | 0.22 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
1738 |
0.46 |
| chr16_91466275_91466861 | 0.22 |
Gm49626 |
predicted gene, 49626 |
1443 |
0.19 |
| chr15_78116860_78117881 | 0.22 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
2336 |
0.22 |
| chr11_101011506_101011900 | 0.18 |
Atp6v0a1 |
ATPase, H+ transporting, lysosomal V0 subunit A1 |
118 |
0.94 |
| chr9_95561747_95561983 | 0.18 |
Paqr9 |
progestin and adipoQ receptor family member IX |
2208 |
0.22 |
| chr9_74977325_74977617 | 0.18 |
Fam214a |
family with sequence similarity 214, member A |
1360 |
0.45 |
| chrX_165326400_165326667 | 0.17 |
Glra2 |
glycine receptor, alpha 2 subunit |
860 |
0.76 |
| chr15_13367879_13368144 | 0.17 |
Gm8238 |
predicted gene 8238 |
50845 |
0.16 |
| chr4_13754294_13755025 | 0.17 |
Runx1t1 |
RUNX1 translocation partner 1 |
3362 |
0.37 |
| chr1_136132801_136134260 | 0.17 |
Kif21b |
kinesin family member 21B |
2076 |
0.19 |
| chr9_10903129_10903698 | 0.16 |
Gm32710 |
predicted gene, 32710 |
932 |
0.44 |
| chr1_164454554_164455430 | 0.16 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1215 |
0.39 |
| chr11_61453075_61454398 | 0.15 |
Rnf112 |
ring finger protein 112 |
183 |
0.92 |
| chr11_62005417_62006894 | 0.15 |
Specc1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
805 |
0.61 |
| chr10_101682665_101683088 | 0.15 |
Mgat4c |
MGAT4 family, member C |
1130 |
0.6 |
| chr15_85675143_85675686 | 0.15 |
Lncppara |
long noncoding RNA near Ppara |
21798 |
0.12 |
| chr6_28827182_28827949 | 0.14 |
Lrrc4 |
leucine rich repeat containing 4 |
2780 |
0.27 |
| chr9_95562029_95562400 | 0.14 |
Paqr9 |
progestin and adipoQ receptor family member IX |
2557 |
0.2 |
| chr4_82589377_82589629 | 0.14 |
Gm11267 |
predicted gene 11267 |
2184 |
0.36 |
| chr10_90577565_90578158 | 0.14 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
869 |
0.72 |
| chr8_86615787_86615938 | 0.13 |
Lonp2 |
lon peptidase 2, peroxisomal |
8181 |
0.17 |
| chr1_146500124_146500296 | 0.13 |
Brinp3 |
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
2472 |
0.28 |
| chr2_97470022_97470462 | 0.13 |
Lrrc4c |
leucine rich repeat containing 4C |
2153 |
0.46 |
| chr9_21196536_21198489 | 0.13 |
Pde4a |
phosphodiesterase 4A, cAMP specific |
807 |
0.45 |
| chr10_77112255_77113959 | 0.13 |
Col18a1 |
collagen, type XVIII, alpha 1 |
598 |
0.73 |
| chr12_71048661_71048812 | 0.13 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
395 |
0.84 |
| chr9_112232861_112233588 | 0.13 |
Arpp21 |
cyclic AMP-regulated phosphoprotein, 21 |
402 |
0.75 |
| chr5_43236032_43236320 | 0.13 |
Gm7854 |
predicted gene 7854 |
821 |
0.45 |
| chr18_31445651_31446131 | 0.13 |
Syt4 |
synaptotagmin IV |
1515 |
0.34 |
| chr3_80798105_80798468 | 0.13 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
4293 |
0.29 |
| chr6_87424489_87424700 | 0.12 |
Bmp10 |
bone morphogenetic protein 10 |
4400 |
0.17 |
| chr11_28582033_28582291 | 0.12 |
Ccdc85a |
coiled-coil domain containing 85A |
1833 |
0.45 |
| chr8_54957960_54958420 | 0.12 |
Gm45263 |
predicted gene 45263 |
1629 |
0.32 |
| chr1_173941256_173941415 | 0.12 |
Ifi203 |
interferon activated gene 203 |
1119 |
0.39 |
| chr8_9767652_9767828 | 0.12 |
Fam155a |
family with sequence similarity 155, member A |
2364 |
0.22 |
| chr13_69736057_69736269 | 0.12 |
Ube2ql1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
3726 |
0.16 |
| chr5_84412572_84412901 | 0.12 |
Epha5 |
Eph receptor A5 |
4070 |
0.29 |
| chr6_55338294_55340060 | 0.12 |
Aqp1 |
aquaporin 1 |
2745 |
0.22 |
| chr16_97168993_97169144 | 0.12 |
Dscam |
DS cell adhesion molecule |
1684 |
0.52 |
| chr3_136673390_136673717 | 0.12 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
2783 |
0.31 |
| chr7_103825389_103825783 | 0.12 |
Hbb-bs |
hemoglobin, beta adult s chain |
2139 |
0.11 |
| chr19_5842574_5845856 | 0.12 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1044 |
0.25 |
| chr4_36948006_36948454 | 0.12 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
2237 |
0.35 |
| chr4_70532291_70532442 | 0.11 |
Megf9 |
multiple EGF-like-domains 9 |
2562 |
0.43 |
| chr1_124042617_124042950 | 0.11 |
Dpp10 |
dipeptidylpeptidase 10 |
2380 |
0.44 |
| chr11_46440995_46441189 | 0.11 |
Med7 |
mediator complex subunit 7 |
1440 |
0.31 |
| chr5_135806693_135807939 | 0.11 |
Srrm3 |
serine/arginine repetitive matrix 3 |
419 |
0.73 |
| chr9_19994680_19994831 | 0.11 |
Olfr864-ps1 |
olfactory receptor 864, pseudogene 1 |
1678 |
0.24 |
| chr14_55054880_55055132 | 0.11 |
Gm20687 |
predicted gene 20687 |
49 |
0.92 |
| chr1_81076133_81076284 | 0.11 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
742 |
0.79 |
| chr6_142962344_142962495 | 0.11 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
1722 |
0.29 |
| chr6_137253502_137253687 | 0.11 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
1129 |
0.6 |
| chr1_173801485_173801725 | 0.11 |
Ifi203-ps |
interferon activated gene 203, pseudogene |
3335 |
0.17 |
| chr14_60381648_60381975 | 0.11 |
Amer2 |
APC membrane recruitment 2 |
3525 |
0.27 |
| chr18_69599939_69600665 | 0.11 |
Tcf4 |
transcription factor 4 |
766 |
0.75 |
| chr11_104234777_104235157 | 0.11 |
Mapt |
microtubule-associated protein tau |
3383 |
0.19 |
| chr12_108605770_108606876 | 0.11 |
Evl |
Ena-vasodilator stimulated phosphoprotein |
557 |
0.74 |
| chr7_80200040_80200211 | 0.11 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
1589 |
0.23 |
| chr18_31442020_31442171 | 0.11 |
Gm26658 |
predicted gene, 26658 |
1777 |
0.3 |
| chr7_61981610_61982350 | 0.11 |
A330076H08Rik |
RIKEN cDNA A330076H08 gene |
323 |
0.52 |
| chr8_46472677_46473081 | 0.11 |
Gm45244 |
predicted gene 45244 |
711 |
0.56 |
| chr16_76318105_76318441 | 0.10 |
Nrip1 |
nuclear receptor interacting protein 1 |
5385 |
0.27 |
| chr14_34675529_34675753 | 0.10 |
Wapl |
WAPL cohesin release factor |
1468 |
0.27 |
| chr7_51630779_51630985 | 0.10 |
Gm45072 |
predicted gene 45072 |
843 |
0.52 |
| chr11_31874060_31874636 | 0.10 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
1073 |
0.55 |
| chr18_43390784_43391261 | 0.10 |
Dpysl3 |
dihydropyrimidinase-like 3 |
2355 |
0.34 |
| chr7_131968717_131968868 | 0.10 |
Gpr26 |
G protein-coupled receptor 26 |
2332 |
0.32 |
| chr2_103958009_103958847 | 0.10 |
Lmo2 |
LIM domain only 2 |
433 |
0.78 |
| chr3_149069217_149069368 | 0.10 |
Gm25127 |
predicted gene, 25127 |
40360 |
0.16 |
| chr10_12611642_12612156 | 0.10 |
Utrn |
utrophin |
2932 |
0.38 |
| chr2_138277835_138280637 | 0.10 |
Btbd3 |
BTB (POZ) domain containing 3 |
743 |
0.81 |
| chr14_108909489_108909909 | 0.10 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
4459 |
0.37 |
| chr9_37527353_37531611 | 0.10 |
Esam |
endothelial cell-specific adhesion molecule |
701 |
0.51 |
| chr3_89522754_89523224 | 0.10 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
2825 |
0.22 |
| chr9_68888905_68889137 | 0.10 |
Rora |
RAR-related orphan receptor alpha |
233718 |
0.02 |
| chr16_92824962_92826063 | 0.10 |
Runx1 |
runt related transcription factor 1 |
266 |
0.94 |
| chr7_87588060_87588319 | 0.10 |
Grm5 |
glutamate receptor, metabotropic 5 |
3791 |
0.35 |
| chr13_42711375_42711526 | 0.10 |
Phactr1 |
phosphatase and actin regulator 1 |
1869 |
0.43 |
| chr3_136672766_136672917 | 0.10 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
2071 |
0.37 |
| chr18_62176067_62177775 | 0.10 |
Adrb2 |
adrenergic receptor, beta 2 |
3038 |
0.24 |
| chr2_134825693_134826026 | 0.10 |
Gm14036 |
predicted gene 14036 |
21910 |
0.2 |
| chr8_46496910_46497239 | 0.10 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
4242 |
0.18 |
| chr6_67034885_67035063 | 0.10 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
1625 |
0.23 |
| chr8_90869910_90870860 | 0.10 |
Gm45640 |
predicted gene 45640 |
6196 |
0.14 |
| chr15_54572723_54573229 | 0.10 |
Mal2 |
mal, T cell differentiation protein 2 |
1784 |
0.45 |
| chr2_94055302_94055453 | 0.10 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
11092 |
0.18 |
| chrX_23283125_23283785 | 0.10 |
Klhl13 |
kelch-like 13 |
1374 |
0.57 |
| chr14_101841859_101842332 | 0.10 |
Lmo7 |
LIM domain only 7 |
1276 |
0.57 |
| chr8_109245493_109246323 | 0.10 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
3958 |
0.33 |
| chr8_61515132_61515409 | 0.10 |
Palld |
palladin, cytoskeletal associated protein |
630 |
0.77 |
| chr15_59079090_59079241 | 0.10 |
Mtss1 |
MTSS I-BAR domain containing 1 |
2824 |
0.32 |
| chrX_144686917_144687223 | 0.09 |
Trpc5 |
transient receptor potential cation channel, subfamily C, member 5 |
933 |
0.55 |
| chr16_42907540_42907820 | 0.09 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
28 |
0.98 |
| chrX_111443823_111443974 | 0.09 |
Rps6ka6 |
ribosomal protein S6 kinase polypeptide 6 |
18741 |
0.28 |
| chr1_79437828_79438003 | 0.09 |
Scg2 |
secretogranin II |
2127 |
0.4 |
| chr8_121730928_121732115 | 0.09 |
Jph3 |
junctophilin 3 |
954 |
0.49 |
| chr13_20474951_20475127 | 0.09 |
Gm32036 |
predicted gene, 32036 |
1549 |
0.33 |
| chr16_46008622_46009173 | 0.09 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
1321 |
0.41 |
| chr3_96181539_96182795 | 0.09 |
Sv2a |
synaptic vesicle glycoprotein 2 a |
1016 |
0.22 |
| chr2_82055274_82055801 | 0.09 |
Zfp804a |
zinc finger protein 804A |
2315 |
0.43 |
| chr3_152866007_152866344 | 0.09 |
Gm16232 |
predicted gene 16232 |
10042 |
0.2 |
| chr10_81426043_81427197 | 0.09 |
Nfic |
nuclear factor I/C |
494 |
0.54 |
| chr11_87762635_87763192 | 0.09 |
Tspoap1 |
TSPO associated protein 1 |
1818 |
0.17 |
| chr11_90196330_90196540 | 0.09 |
Gm45883 |
predicted gene 45883 |
8260 |
0.16 |
| chr5_66678971_66679451 | 0.09 |
Uchl1 |
ubiquitin carboxy-terminal hydrolase L1 |
2319 |
0.2 |
| chr13_117022581_117022756 | 0.09 |
Parp8 |
poly (ADP-ribose) polymerase family, member 8 |
2605 |
0.3 |
| chr19_38265533_38266016 | 0.09 |
Lgi1 |
leucine-rich repeat LGI family, member 1 |
364 |
0.85 |
| chr1_3667888_3668100 | 0.09 |
Xkr4 |
X-linked Kx blood group related 4 |
3504 |
0.21 |
| chr15_78404813_78404964 | 0.09 |
Tst |
thiosulfate sulfurtransferase, mitochondrial |
964 |
0.32 |
| chr12_5372599_5372750 | 0.09 |
Klhl29 |
kelch-like 29 |
3008 |
0.28 |
| chr11_115511512_115512231 | 0.09 |
Jpt1 |
Jupiter microtubule associated homolog 1 |
2245 |
0.14 |
| chr1_70725543_70726581 | 0.09 |
Vwc2l |
von Willebrand factor C domain-containing protein 2-like |
139 |
0.98 |
| chr13_59819810_59820064 | 0.09 |
Tut7 |
terminal uridylyl transferase 7 |
2651 |
0.15 |
| chr2_23562980_23563209 | 0.09 |
Spopl |
speckle-type BTB/POZ protein-like |
8884 |
0.24 |
| chr16_77236514_77236677 | 0.09 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
276 |
0.93 |
| chr18_56922535_56922923 | 0.09 |
Marchf3 |
membrane associated ring-CH-type finger 3 |
2786 |
0.27 |
| chr12_56253431_56254088 | 0.09 |
Gm19990 |
predicted gene, 19990 |
516 |
0.47 |
| chr7_122673354_122673569 | 0.09 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
2043 |
0.32 |
| chr6_120819578_120819729 | 0.09 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
1254 |
0.37 |
| chr15_36275796_36275947 | 0.09 |
Rnf19a |
ring finger protein 19A |
7227 |
0.13 |
| chr5_151188598_151188749 | 0.09 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
1477 |
0.48 |
| chr14_64575803_64576191 | 0.09 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
11334 |
0.15 |
| chr6_24047175_24047352 | 0.09 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
61308 |
0.12 |
| chr4_32862939_32864770 | 0.09 |
Ankrd6 |
ankyrin repeat domain 6 |
3171 |
0.24 |
| chr8_121904943_121905414 | 0.09 |
Slc7a5 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
2516 |
0.15 |
| chr2_148404766_148404943 | 0.09 |
Thbd |
thrombomodulin |
3334 |
0.2 |
| chr10_111166520_111166791 | 0.09 |
Osbpl8 |
oxysterol binding protein-like 8 |
1853 |
0.27 |
| chr3_94412462_94413013 | 0.09 |
1700040D17Rik |
RIKEN cDNA 1700040D17 gene |
182 |
0.67 |
| chr15_78571734_78572724 | 0.09 |
Rac2 |
Rac family small GTPase 2 |
552 |
0.62 |
| chr1_75277211_75278430 | 0.09 |
Resp18 |
regulated endocrine-specific protein 18 |
464 |
0.62 |
| chr4_138251495_138252773 | 0.09 |
Sh2d5 |
SH2 domain containing 5 |
1670 |
0.22 |
| chr19_26751454_26751605 | 0.09 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
407 |
0.88 |
| chr19_8840785_8841292 | 0.09 |
Bscl2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
503 |
0.44 |
| chr7_126823319_126824529 | 0.09 |
Fam57b |
family with sequence similarity 57, member B |
621 |
0.41 |
| chr5_9721072_9721322 | 0.09 |
Grm3 |
glutamate receptor, metabotropic 3 |
3973 |
0.27 |
| chr8_93814307_93815014 | 0.09 |
4930488L21Rik |
RIKEN cDNA 4930488L21 gene |
938 |
0.54 |
| chrX_85870254_85870648 | 0.09 |
5430427O19Rik |
RIKEN cDNA 5430427O19 gene |
97 |
0.97 |
| chr1_162898137_162898420 | 0.09 |
Fmo2 |
flavin containing monooxygenase 2 |
206 |
0.94 |
| chr2_60960139_60960473 | 0.09 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
2886 |
0.3 |
| chr2_24385363_24386049 | 0.09 |
Psd4 |
pleckstrin and Sec7 domain containing 4 |
353 |
0.84 |
| chr5_150261018_150262108 | 0.09 |
Fry |
FRY microtubule binding protein |
1796 |
0.34 |
| chr6_67228511_67229348 | 0.09 |
Gm44052 |
predicted gene, 44052 |
6385 |
0.13 |
| chr1_172501767_172503923 | 0.09 |
Tagln2 |
transgelin 2 |
1593 |
0.22 |
| chr11_15096657_15096828 | 0.09 |
Gm12009 |
predicted gene 12009 |
47584 |
0.2 |
| chr3_51636519_51637197 | 0.08 |
Gm38247 |
predicted gene, 38247 |
70 |
0.96 |
| chr2_6870837_6870988 | 0.08 |
Celf2 |
CUGBP, Elav-like family member 2 |
1060 |
0.55 |
| chr1_38012822_38013138 | 0.08 |
Eif5b |
eukaryotic translation initiation factor 5B |
14851 |
0.14 |
| chr1_140182842_140183316 | 0.08 |
Cfh |
complement component factor h |
201 |
0.96 |
| chr11_20331076_20331227 | 0.08 |
Slc1a4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
1533 |
0.37 |
| chr15_37457581_37457955 | 0.08 |
Ncald |
neurocalcin delta |
773 |
0.56 |
| chrX_167207304_167207608 | 0.08 |
Tmsb4x |
thymosin, beta 4, X chromosome |
1371 |
0.45 |
| chr11_43834010_43835381 | 0.08 |
Adra1b |
adrenergic receptor, alpha 1b |
1637 |
0.47 |
| chr10_33256172_33256374 | 0.08 |
D830005E20Rik |
RIKEN cDNA D830005E20 gene |
84 |
0.97 |
| chr1_115687470_115687621 | 0.08 |
Cntnap5a |
contactin associated protein-like 5A |
2789 |
0.31 |
| chr17_44809334_44809504 | 0.08 |
Runx2 |
runt related transcription factor 2 |
4807 |
0.23 |
| chr15_98674837_98676155 | 0.08 |
Rnd1 |
Rho family GTPase 1 |
1962 |
0.18 |
| chr17_91085493_91086001 | 0.08 |
Gm47307 |
predicted gene, 47307 |
2659 |
0.21 |
| chr3_14890853_14891237 | 0.08 |
Car2 |
carbonic anhydrase 2 |
4406 |
0.22 |
| chr14_101842332_101842898 | 0.08 |
Lmo7 |
LIM domain only 7 |
1796 |
0.46 |
| chr16_46008308_46008480 | 0.08 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
1824 |
0.32 |
| chr13_90844060_90844211 | 0.08 |
Gm18518 |
predicted gene, 18518 |
23471 |
0.19 |
| chr6_39874717_39875333 | 0.08 |
Tmem178b |
transmembrane protein 178B |
1954 |
0.27 |
| chrX_93430021_93430176 | 0.08 |
Pola1 |
polymerase (DNA directed), alpha 1 |
60848 |
0.12 |
| chr13_116298582_116298733 | 0.08 |
Isl1 |
ISL1 transcription factor, LIM/homeodomain |
4694 |
0.26 |
| chr14_27876139_27876947 | 0.08 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
129 |
0.75 |
| chr3_4800881_4801176 | 0.08 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
1535 |
0.47 |
| chr16_20098725_20098876 | 0.08 |
Klhl24 |
kelch-like 24 |
1208 |
0.49 |
| chr13_116214509_116214705 | 0.08 |
Rpl34-ps2 |
ribosomal protein L34, pseudogene 2 |
60826 |
0.13 |
| chr5_146232824_146233169 | 0.08 |
Cdk8 |
cyclin-dependent kinase 8 |
1611 |
0.25 |
| chr10_96623801_96623952 | 0.08 |
Btg1 |
BTG anti-proliferation factor 1 |
6322 |
0.22 |
| chr16_31423409_31423642 | 0.08 |
Bdh1 |
3-hydroxybutyrate dehydrogenase, type 1 |
1245 |
0.34 |
| chr12_117805814_117806560 | 0.08 |
Cdca7l |
cell division cycle associated 7 like |
1898 |
0.37 |
| chr16_36366547_36366698 | 0.08 |
Gm15845 |
predicted gene 15845 |
448 |
0.53 |
| chrX_95194864_95195015 | 0.08 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
1513 |
0.42 |
| chr10_63977869_63978469 | 0.08 |
Gm10118 |
predicted gene 10118 |
50735 |
0.17 |
| chr12_113426567_113426898 | 0.08 |
Gm22175 |
predicted gene, 22175 |
1505 |
0.16 |
| chr10_42273694_42273969 | 0.08 |
Foxo3 |
forkhead box O3 |
2865 |
0.34 |
| chr2_103971594_103971745 | 0.08 |
Lmo2 |
LIM domain only 2 |
490 |
0.74 |
| chr14_70618232_70619317 | 0.08 |
Dmtn |
dematin actin binding protein |
229 |
0.88 |
| chrX_88113433_88114223 | 0.08 |
Il1rapl1 |
interleukin 1 receptor accessory protein-like 1 |
1817 |
0.46 |
| chr1_130721109_130721366 | 0.08 |
Yod1 |
YOD1 deubiquitinase |
3910 |
0.11 |
| chr14_27380191_27380512 | 0.08 |
Gm23633 |
predicted gene, 23633 |
499 |
0.8 |
| chr7_127767459_127768302 | 0.08 |
Orai3 |
ORAI calcium release-activated calcium modulator 3 |
1935 |
0.14 |
| chr11_34047188_34048526 | 0.08 |
Lcp2 |
lymphocyte cytosolic protein 2 |
689 |
0.72 |
| chr16_54635653_54635804 | 0.08 |
Gm23180 |
predicted gene, 23180 |
7300 |
0.29 |
| chr18_56756025_56756589 | 0.08 |
Gm15345 |
predicted gene 15345 |
13410 |
0.18 |
| chr16_79109471_79109622 | 0.08 |
Tmprss15 |
transmembrane protease, serine 15 |
18449 |
0.28 |
| chr12_86424576_86424958 | 0.08 |
Esrrb |
estrogen related receptor, beta |
2884 |
0.33 |
| chr2_65848318_65848473 | 0.08 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
2540 |
0.31 |
| chr18_32539971_32540198 | 0.08 |
Gypc |
glycophorin C |
4666 |
0.22 |
| chr10_81497570_81499812 | 0.08 |
S1pr4 |
sphingosine-1-phosphate receptor 4 |
1441 |
0.16 |
| chr12_113141740_113143605 | 0.08 |
Crip2 |
cysteine rich protein 2 |
136 |
0.92 |
| chr9_121403057_121404479 | 0.08 |
Trak1 |
trafficking protein, kinesin binding 1 |
290 |
0.91 |
| chrX_128439905_128440056 | 0.08 |
Gm38390 |
predicted gene, 38390 |
64073 |
0.12 |
| chr7_99537223_99537701 | 0.08 |
Arrb1 |
arrestin, beta 1 |
1758 |
0.23 |
| chr11_37083444_37083595 | 0.08 |
Tenm2 |
teneurin transmembrane protein 2 |
139353 |
0.05 |
| chr6_83171941_83172736 | 0.08 |
Gm15624 |
predicted gene 15624 |
216 |
0.83 |
| chr3_88501925_88503550 | 0.08 |
Lmna |
lamin A |
570 |
0.53 |
| chr1_182281173_182281731 | 0.08 |
Degs1 |
delta(4)-desaturase, sphingolipid 1 |
772 |
0.62 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0098828 | modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.0 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.0 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0052714 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0016937 | short-branched-chain-acyl-CoA dehydrogenase activity(GO:0016937) |
| 0.0 | 0.1 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |