Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
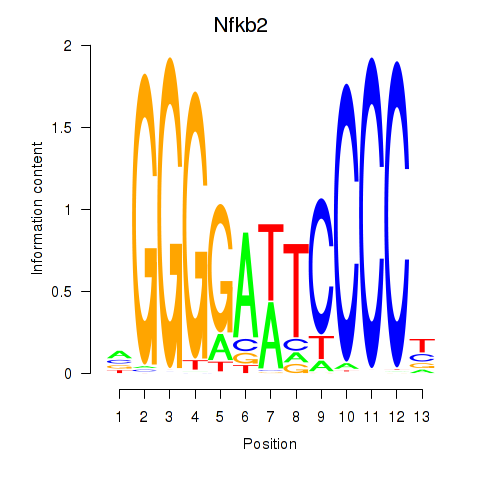
Results for Nfkb2
Z-value: 0.68
Transcription factors associated with Nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfkb2
|
ENSMUSG00000025225.8 | nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
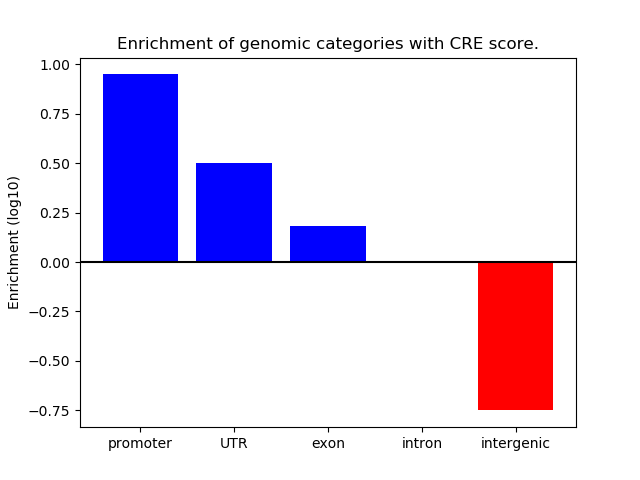
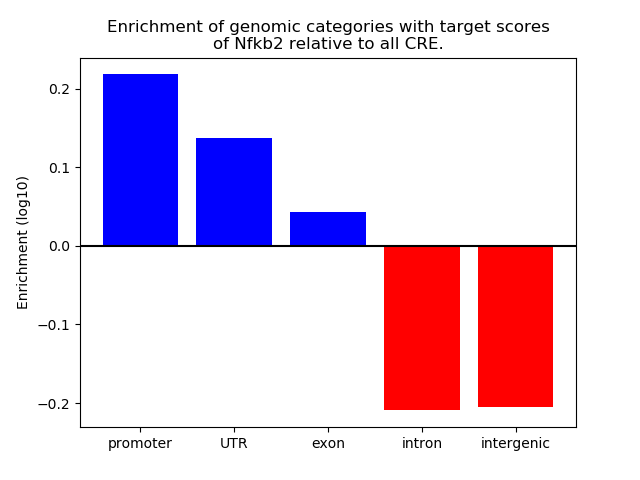
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_46308370_46309175 | Nfkb2 | 2651 | 0.121170 | 0.37 | 3.8e-03 | Click! |
| chr19_46309210_46309553 | Nfkb2 | 3260 | 0.107330 | 0.27 | 3.5e-02 | Click! |
| chr19_46309835_46310023 | Nfkb2 | 3808 | 0.099537 | 0.18 | 1.6e-01 | Click! |
| chr19_46304094_46304245 | Nfkb2 | 151 | 0.883432 | -0.16 | 2.2e-01 | Click! |
| chr19_46308192_46308343 | Nfkb2 | 2146 | 0.141029 | 0.14 | 2.8e-01 | Click! |
Activity of the Nfkb2 motif across conditions
Conditions sorted by the z-value of the Nfkb2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
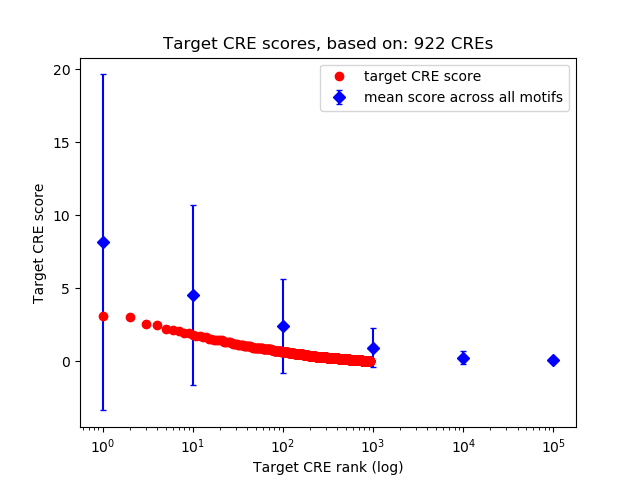
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_70119024_70120981 | 3.07 |
Ncan |
neurocan |
871 |
0.35 |
| chr2_143546820_143547517 | 3.02 |
Pcsk2os1 |
proprotein convertase subtilisin/kexin type 2, opposite strand 1 |
669 |
0.53 |
| chr14_93888463_93888979 | 2.58 |
Pcdh9 |
protocadherin 9 |
11 |
0.99 |
| chr2_65929929_65930575 | 2.51 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
115 |
0.97 |
| chr17_25567606_25567996 | 2.20 |
Sox8 |
SRY (sex determining region Y)-box 8 |
2413 |
0.13 |
| chr15_12739006_12740203 | 2.13 |
Pdzd2 |
PDZ domain containing 2 |
320 |
0.86 |
| chr13_105444000_105445296 | 2.05 |
Htr1a |
5-hydroxytryptamine (serotonin) receptor 1A |
1009 |
0.69 |
| chr14_67236008_67239452 | 1.95 |
Ebf2 |
early B cell factor 2 |
3086 |
0.21 |
| chr9_73967774_73969221 | 1.93 |
Unc13c |
unc-13 homolog C |
469 |
0.88 |
| chr5_49284738_49286021 | 1.80 |
Kcnip4 |
Kv channel interacting protein 4 |
280 |
0.92 |
| chr2_38354292_38355594 | 1.75 |
Lhx2 |
LIM homeobox protein 2 |
1141 |
0.41 |
| chr1_178530778_178530954 | 1.70 |
Kif26b |
kinesin family member 26B |
1741 |
0.44 |
| chr5_142961791_142962040 | 1.68 |
Fscn1 |
fascin actin-bundling protein 1 |
1038 |
0.49 |
| chr15_59319527_59320112 | 1.67 |
Sqle |
squalene epoxidase |
4712 |
0.18 |
| chrX_135210129_135210918 | 1.51 |
Tceal6 |
transcription elongation factor A (SII)-like 6 |
164 |
0.93 |
| chr13_112289274_112289896 | 1.50 |
Ankrd55 |
ankyrin repeat domain 55 |
765 |
0.56 |
| chr2_27027194_27027995 | 1.49 |
Slc2a6 |
solute carrier family 2 (facilitated glucose transporter), member 6 |
315 |
0.79 |
| chr1_124045018_124046369 | 1.48 |
Dpp10 |
dipeptidylpeptidase 10 |
134 |
0.98 |
| chr3_89522754_89523224 | 1.44 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
2825 |
0.22 |
| chr11_96987508_96988742 | 1.43 |
Sp2 |
Sp2 transcription factor |
5166 |
0.1 |
| chr10_114800824_114801254 | 1.43 |
Trhde |
TRH-degrading enzyme |
331 |
0.73 |
| chr17_57148993_57149872 | 1.35 |
Cd70 |
CD70 antigen |
345 |
0.79 |
| chr12_79130802_79131751 | 1.34 |
Arg2 |
arginase type II |
499 |
0.64 |
| chr18_69347722_69348433 | 1.31 |
Tcf4 |
transcription factor 4 |
179 |
0.97 |
| chr8_83902724_83903561 | 1.30 |
Adgrl1 |
adhesion G protein-coupled receptor L1 |
2407 |
0.18 |
| chr7_79507205_79507895 | 1.29 |
Mir9-3 |
microRNA 9-3 |
2286 |
0.14 |
| chr15_78717784_78718525 | 1.25 |
Elfn2 |
leucine rich repeat and fibronectin type III, extracellular 2 |
41 |
0.97 |
| chr10_80179768_80180422 | 1.19 |
Efna2 |
ephrin A2 |
613 |
0.51 |
| chr15_102959654_102960750 | 1.19 |
Hoxc11 |
homeobox C11 |
5775 |
0.08 |
| chr6_110645148_110646464 | 1.17 |
Gm20387 |
predicted gene 20387 |
110 |
0.67 |
| chr5_28465805_28466194 | 1.15 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
985 |
0.44 |
| chr11_11115362_11116603 | 1.15 |
Vwc2 |
von Willebrand factor C domain containing 2 |
131 |
0.97 |
| chr1_74802832_74804032 | 1.11 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
10069 |
0.11 |
| chr4_140647825_140648405 | 1.10 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
635 |
0.68 |
| chr15_98951099_98951690 | 1.09 |
Gm49450 |
predicted gene, 49450 |
2157 |
0.13 |
| chr14_88469423_88470289 | 1.09 |
Pcdh20 |
protocadherin 20 |
1490 |
0.47 |
| chr11_93100564_93101210 | 1.07 |
Car10 |
carbonic anhydrase 10 |
1597 |
0.56 |
| chrX_102069269_102069420 | 1.07 |
Rtl5 |
retrotransposon Gag like 5 |
1744 |
0.27 |
| chr11_96297868_96298678 | 1.06 |
Hoxb6 |
homeobox B6 |
898 |
0.3 |
| chr7_131966504_131967699 | 1.06 |
Gpr26 |
G protein-coupled receptor 26 |
641 |
0.75 |
| chr18_83635368_83635622 | 1.05 |
Gm31621 |
predicted gene, 31621 |
2431 |
0.28 |
| chr7_45869909_45870408 | 1.03 |
Grin2d |
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
271 |
0.77 |
| chr1_19104045_19104891 | 1.03 |
Gm15825 |
predicted gene 15825 |
372 |
0.8 |
| chr5_90758918_90760166 | 1.00 |
Cxcl5 |
chemokine (C-X-C motif) ligand 5 |
164 |
0.91 |
| chr11_102081725_102082491 | 0.99 |
Mpp2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
311 |
0.79 |
| chr7_25007594_25007745 | 0.95 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
1711 |
0.22 |
| chr14_58150040_58150734 | 0.94 |
Gm17109 |
predicted gene 17109 |
44258 |
0.14 |
| chr11_69800320_69800731 | 0.93 |
Fgf11 |
fibroblast growth factor 11 |
1191 |
0.19 |
| chr3_88458101_88459325 | 0.92 |
Sema4a |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
163 |
0.88 |
| chr4_154649328_154649479 | 0.92 |
Gm27202 |
predicted gene 27202 |
3702 |
0.15 |
| chr7_74012361_74013617 | 0.92 |
St8sia2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
685 |
0.71 |
| chr15_73668219_73668370 | 0.92 |
1700010B13Rik |
RIKEN cDNA 1700010B13 gene |
22420 |
0.14 |
| chr8_126140788_126141154 | 0.91 |
Slc35f3 |
solute carrier family 35, member F3 |
1589 |
0.49 |
| chr2_18056600_18056751 | 0.91 |
1810059C17Rik |
RIKEN cDNA 1810059C17 gene |
62 |
0.92 |
| chr2_152951095_152951539 | 0.90 |
Dusp15 |
dual specificity phosphatase-like 15 |
139 |
0.93 |
| chr19_16135060_16135656 | 0.89 |
Gnaq |
guanine nucleotide binding protein, alpha q polypeptide |
2062 |
0.35 |
| chr11_34315414_34316667 | 0.89 |
Insyn2b |
inhibitory synaptic factor family member 2B |
1218 |
0.45 |
| chr5_90902965_90904167 | 0.88 |
Cxcl2 |
chemokine (C-X-C motif) ligand 2 |
305 |
0.84 |
| chr11_98980727_98980878 | 0.88 |
Gjd3 |
gap junction protein, delta 3 |
2214 |
0.17 |
| chr9_13301218_13302233 | 0.87 |
Maml2 |
mastermind like transcriptional coactivator 2 |
3768 |
0.19 |
| chr1_177640225_177640394 | 0.87 |
2310043L19Rik |
RIKEN cDNA 2310043L19 gene |
2634 |
0.25 |
| chr2_163602380_163602998 | 0.87 |
Ttpal |
tocopherol (alpha) transfer protein-like |
126 |
0.95 |
| chr6_55455310_55456012 | 0.86 |
Adcyap1r1 |
adenylate cyclase activating polypeptide 1 receptor 1 |
3484 |
0.26 |
| chr3_66976883_66977353 | 0.86 |
Shox2 |
short stature homeobox 2 |
1329 |
0.41 |
| chr8_26848235_26848453 | 0.84 |
2310008N11Rik |
RIKEN cDNA 2310008N11 gene |
6345 |
0.21 |
| chr1_5018491_5020565 | 0.84 |
Rgs20 |
regulator of G-protein signaling 20 |
11 |
0.98 |
| chr3_66219854_66220300 | 0.83 |
Ptx3 |
pentraxin related gene |
167 |
0.95 |
| chr2_33626557_33626708 | 0.83 |
Lmx1b |
LIM homeobox transcription factor 1 beta |
5651 |
0.16 |
| chr2_33593442_33593593 | 0.83 |
Gm38011 |
predicted gene, 38011 |
20881 |
0.15 |
| chr7_126848132_126848884 | 0.83 |
Doc2a |
double C2, alpha |
253 |
0.76 |
| chr11_58866994_58868023 | 0.83 |
2810021J22Rik |
RIKEN cDNA 2810021J22 gene |
241 |
0.79 |
| chr12_98577628_98578516 | 0.83 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
238 |
0.9 |
| chr5_96373327_96373478 | 0.81 |
Fras1 |
Fraser extracellular matrix complex subunit 1 |
553 |
0.83 |
| chr5_113490881_113491656 | 0.81 |
Wscd2 |
WSC domain containing 2 |
516 |
0.83 |
| chr1_12991432_12992703 | 0.80 |
Slco5a1 |
solute carrier organic anion transporter family, member 5A1 |
583 |
0.78 |
| chr10_18341675_18342214 | 0.78 |
Gm18942 |
predicted gene, 18942 |
4703 |
0.22 |
| chr10_81251490_81251641 | 0.76 |
Matk |
megakaryocyte-associated tyrosine kinase |
1370 |
0.17 |
| chr12_89811728_89812295 | 0.74 |
Nrxn3 |
neurexin III |
397 |
0.92 |
| chr7_43489310_43490670 | 0.74 |
Iglon5 |
IgLON family member 5 |
85 |
0.92 |
| chr1_119051596_119052429 | 0.73 |
Gli2 |
GLI-Kruppel family member GLI2 |
1327 |
0.47 |
| chr11_44632092_44632243 | 0.73 |
Ebf1 |
early B cell factor 1 |
10406 |
0.17 |
| chr11_116809628_116809779 | 0.72 |
Mxra7 |
matrix-remodelling associated 7 |
18300 |
0.1 |
| chr17_34038427_34040461 | 0.72 |
Col11a2 |
collagen, type XI, alpha 2 |
7 |
0.9 |
| chr16_97168581_97168970 | 0.72 |
Dscam |
DS cell adhesion molecule |
1977 |
0.47 |
| chr1_172482102_172483290 | 0.72 |
Igsf9 |
immunoglobulin superfamily, member 9 |
382 |
0.75 |
| chr7_45795764_45796042 | 0.72 |
Lmtk3 |
lemur tyrosine kinase 3 |
8155 |
0.07 |
| chr1_191224508_191225332 | 0.71 |
D730003I15Rik |
RIKEN cDNA D730003I15 gene |
446 |
0.76 |
| chr3_3308823_3309359 | 0.70 |
Gm8747 |
predicted gene 8747 |
42064 |
0.2 |
| chr19_10356843_10356994 | 0.70 |
Syt7 |
synaptotagmin VII |
32172 |
0.12 |
| chr15_103519597_103520253 | 0.69 |
Pde1b |
phosphodiesterase 1B, Ca2+-calmodulin dependent |
3793 |
0.17 |
| chr2_102450383_102451888 | 0.69 |
Fjx1 |
four jointed box 1 |
1364 |
0.51 |
| chr4_126277866_126278017 | 0.69 |
Col8a2 |
collagen, type VIII, alpha 2 |
8852 |
0.12 |
| chr15_103105344_103105529 | 0.69 |
Gm28265 |
predicted gene 28265 |
834 |
0.41 |
| chr2_150361232_150361675 | 0.68 |
3300002I08Rik |
RIKEN cDNA 3300002I08 gene |
1280 |
0.35 |
| chr15_84301565_84301928 | 0.68 |
Gm22890 |
predicted gene, 22890 |
21606 |
0.13 |
| chr10_59221970_59222690 | 0.67 |
Sowahc |
sosondowah ankyrin repeat domain family member C |
377 |
0.54 |
| chr6_86051805_86052622 | 0.66 |
Add2 |
adducin 2 (beta) |
35 |
0.96 |
| chr2_77816072_77817449 | 0.66 |
Zfp385b |
zinc finger protein 385B |
56 |
0.98 |
| chr11_118568846_118570341 | 0.66 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
317 |
0.91 |
| chr9_119092635_119093830 | 0.66 |
Plcd1 |
phospholipase C, delta 1 |
270 |
0.87 |
| chr15_81696545_81697665 | 0.65 |
Chadl |
chondroadherin-like |
182 |
0.9 |
| chr4_102087325_102088593 | 0.64 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
218 |
0.95 |
| chr15_102980564_102980932 | 0.63 |
Hoxc9 |
homeobox C9 |
3716 |
0.09 |
| chr8_111742802_111743840 | 0.63 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
488 |
0.8 |
| chr18_42401018_42401905 | 0.63 |
Pou4f3 |
POU domain, class 4, transcription factor 3 |
6386 |
0.2 |
| chr10_127187269_127187420 | 0.63 |
Arhgef25 |
Rho guanine nucleotide exchange factor (GEF) 25 |
121 |
0.91 |
| chr2_27122326_27122477 | 0.62 |
Adamtsl2 |
ADAMTS-like 2 |
17738 |
0.11 |
| chr19_55749292_55750940 | 0.62 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
7271 |
0.3 |
| chr9_41474743_41475732 | 0.62 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
177 |
0.72 |
| chr11_3910794_3911677 | 0.62 |
Slc35e4 |
solute carrier family 35, member E4 |
3429 |
0.13 |
| chr13_42709240_42709536 | 0.61 |
Phactr1 |
phosphatase and actin regulator 1 |
155 |
0.97 |
| chr4_129966249_129966400 | 0.61 |
1700003M07Rik |
RIKEN cDNA 1700003M07 gene |
5516 |
0.14 |
| chr17_28144735_28145006 | 0.61 |
Scube3 |
signal peptide, CUB domain, EGF-like 3 |
2344 |
0.18 |
| chr12_72844062_72844595 | 0.60 |
Gm33785 |
predicted gene, 33785 |
26616 |
0.16 |
| chr11_97627602_97627753 | 0.60 |
Epop |
elongin BC and polycomb repressive complex 2 associated protein |
2025 |
0.18 |
| chr2_25580658_25582212 | 0.60 |
Ajm1 |
apical junction component 1 |
293 |
0.72 |
| chr5_125158383_125159717 | 0.59 |
Ncor2 |
nuclear receptor co-repressor 2 |
20003 |
0.19 |
| chrX_73411302_73411931 | 0.59 |
Zfp92 |
zinc finger protein 92 |
56 |
0.66 |
| chr19_59468752_59469845 | 0.58 |
Emx2 |
empty spiracles homeobox 2 |
6496 |
0.17 |
| chr7_45417677_45418655 | 0.58 |
AC151602.1 |
luteinizing hormone beta |
187 |
0.8 |
| chr1_129275124_129275766 | 0.58 |
Thsd7b |
thrombospondin, type I, domain containing 7B |
1960 |
0.39 |
| chr10_94311604_94312850 | 0.58 |
Tmcc3 |
transmembrane and coiled coil domains 3 |
90 |
0.97 |
| chr5_67633750_67634259 | 0.57 |
Gm42735 |
predicted gene 42735 |
1304 |
0.3 |
| chr11_5255980_5256131 | 0.57 |
Gm25142 |
predicted gene, 25142 |
4205 |
0.19 |
| chr2_25460160_25461436 | 0.57 |
Paxx |
non-homologous end joining factor |
129 |
0.89 |
| chr11_109297097_109298424 | 0.57 |
Rgs9 |
regulator of G-protein signaling 9 |
306 |
0.9 |
| chr7_46099780_46100565 | 0.56 |
Kcnj11 |
potassium inwardly rectifying channel, subfamily J, member 11 |
65 |
0.95 |
| chr19_5093217_5093838 | 0.56 |
Yif1a |
Yip1 interacting factor homolog A (S. cerevisiae) |
4599 |
0.07 |
| chr7_101419184_101419783 | 0.56 |
Pde2a |
phosphodiesterase 2A, cGMP-stimulated |
2208 |
0.21 |
| chr16_91012571_91012722 | 0.55 |
4930404I05Rik |
RIKEN cDNA 4930404I05 gene |
1075 |
0.29 |
| chr10_102160210_102160629 | 0.55 |
Mgat4c |
MGAT4 family, member C |
1340 |
0.6 |
| chr10_127619791_127621096 | 0.55 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
479 |
0.65 |
| chr9_106456260_106457377 | 0.54 |
Pcbp4 |
poly(rC) binding protein 4 |
721 |
0.42 |
| chr4_121039154_121040672 | 0.53 |
Col9a2 |
collagen, type IX, alpha 2 |
165 |
0.92 |
| chr18_38216257_38217428 | 0.53 |
2010320O07Rik |
RIKEN cDNA 2010320O07 gene |
3138 |
0.16 |
| chr2_32311532_32312331 | 0.53 |
Dnm1 |
dynamin 1 |
958 |
0.33 |
| chr11_35120980_35122375 | 0.52 |
Slit3 |
slit guidance ligand 3 |
453 |
0.89 |
| chr5_117087628_117088305 | 0.52 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
10845 |
0.15 |
| chr2_152907676_152907827 | 0.52 |
Mylk2 |
myosin, light polypeptide kinase 2, skeletal muscle |
3601 |
0.15 |
| chr11_98950065_98950283 | 0.52 |
Gm22061 |
predicted gene, 22061 |
3730 |
0.13 |
| chr6_37297951_37298469 | 0.52 |
Dgki |
diacylglycerol kinase, iota |
1427 |
0.53 |
| chr5_121823215_121823366 | 0.52 |
Mir7031 |
microRNA 7031 |
4017 |
0.12 |
| chr2_35619545_35620730 | 0.52 |
Dab2ip |
disabled 2 interacting protein |
1844 |
0.38 |
| chr2_27460067_27460218 | 0.52 |
Brd3 |
bromodomain containing 3 |
3974 |
0.16 |
| chr2_32759044_32759195 | 0.51 |
Tor2a |
torsin family 2, member A |
1176 |
0.25 |
| chr4_155252152_155252689 | 0.51 |
Faap20 |
Fanconi anemia core complex associated protein 20 |
1724 |
0.32 |
| chr16_45723390_45724960 | 0.50 |
Tagln3 |
transgelin 3 |
433 |
0.77 |
| chr5_99726980_99727219 | 0.50 |
Rasgef1b |
RasGEF domain family, member 1B |
1713 |
0.32 |
| chr16_91229951_91230102 | 0.50 |
Olig2 |
oligodendrocyte transcription factor 2 |
4569 |
0.14 |
| chr16_77236731_77239778 | 0.50 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
1935 |
0.4 |
| chr8_105699776_105699927 | 0.50 |
Acd |
adrenocortical dysplasia |
30 |
0.93 |
| chr8_24296964_24297158 | 0.49 |
Gm45164 |
predicted gene 45164 |
62375 |
0.11 |
| chr10_127488582_127488733 | 0.49 |
R3hdm2 |
R3H domain containing 2 |
3175 |
0.14 |
| chr11_96697035_96697186 | 0.49 |
Skap1 |
src family associated phosphoprotein 1 |
11125 |
0.17 |
| chrX_7863281_7863432 | 0.49 |
Otud5 |
OTU domain containing 5 |
7629 |
0.08 |
| chr1_34801752_34802642 | 0.49 |
Arhgef4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
403 |
0.78 |
| chr7_80320493_80320644 | 0.48 |
Rccd1 |
RCC1 domain containing 1 |
471 |
0.66 |
| chr5_143293458_143294037 | 0.48 |
Zfp853 |
zinc finger protein 853 |
124 |
0.91 |
| chr12_12938033_12939196 | 0.48 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
2002 |
0.23 |
| chr5_69340137_69340288 | 0.48 |
Gm24368 |
predicted gene, 24368 |
571 |
0.63 |
| chr15_101253842_101254216 | 0.48 |
Nr4a1 |
nuclear receptor subfamily 4, group A, member 1 |
240 |
0.86 |
| chr12_70830034_70830209 | 0.47 |
Frmd6 |
FERM domain containing 6 |
4432 |
0.2 |
| chrX_20290867_20291587 | 0.47 |
Slc9a7 |
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
543 |
0.77 |
| chr9_37783109_37783351 | 0.47 |
Olfr875 |
olfactory receptor 875 |
10593 |
0.1 |
| chr15_84444030_84444699 | 0.47 |
Shisal1 |
shisa like 1 |
667 |
0.75 |
| chr15_103005378_103006088 | 0.46 |
Hoxc6 |
homeobox C6 |
3840 |
0.1 |
| chr4_134496639_134497966 | 0.46 |
Paqr7 |
progestin and adipoQ receptor family member VII |
261 |
0.84 |
| chr2_164961062_164961920 | 0.46 |
Slc12a5 |
solute carrier family 12, member 5 |
645 |
0.6 |
| chr17_56466771_56466941 | 0.46 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
4640 |
0.17 |
| chr1_69105258_69106691 | 0.46 |
Gm16076 |
predicted gene 16076 |
766 |
0.61 |
| chr11_95057837_95057988 | 0.46 |
Itga3 |
integrin alpha 3 |
1717 |
0.25 |
| chr9_102722581_102723136 | 0.45 |
Amotl2 |
angiomotin-like 2 |
324 |
0.83 |
| chr4_127235958_127237081 | 0.45 |
Smim12 |
small integral membrane protein 12 |
7265 |
0.16 |
| chr1_133049808_133049959 | 0.45 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
3871 |
0.17 |
| chr19_46063703_46063854 | 0.45 |
Pprc1 |
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
4056 |
0.15 |
| chr4_28816938_28817154 | 0.44 |
Epha7 |
Eph receptor A7 |
1617 |
0.37 |
| chr3_45432245_45432549 | 0.44 |
Gm2136 |
predicted gene 2136 |
5983 |
0.17 |
| chr3_10329449_10329600 | 0.44 |
Impa1 |
inositol (myo)-1(or 4)-monophosphatase 1 |
1608 |
0.22 |
| chr6_114133449_114133914 | 0.44 |
Slc6a11 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
2262 |
0.31 |
| chr2_3419168_3419605 | 0.44 |
Meig1 |
meiosis expressed gene 1 |
257 |
0.87 |
| chr9_55326922_55327600 | 0.43 |
Tmem266 |
transmembrane protein 266 |
312 |
0.58 |
| chr17_26953045_26954307 | 0.43 |
Syngap1 |
synaptic Ras GTPase activating protein 1 homolog (rat) |
790 |
0.36 |
| chr14_14345697_14346878 | 0.43 |
Il3ra |
interleukin 3 receptor, alpha chain |
12 |
0.94 |
| chr2_102552573_102552724 | 0.42 |
Pamr1 |
peptidase domain containing associated with muscle regeneration 1 |
2499 |
0.38 |
| chr2_72516060_72516494 | 0.41 |
Rps13-ps8 |
ribosmal protein S13, pseudogene 8 |
28935 |
0.16 |
| chr13_97248203_97248451 | 0.41 |
Enc1 |
ectodermal-neural cortex 1 |
7222 |
0.17 |
| chr13_109926306_109927455 | 0.41 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
36 |
0.98 |
| chr2_164388819_164390055 | 0.41 |
Slpi |
secretory leukocyte peptidase inhibitor |
342 |
0.77 |
| chrX_36111786_36112920 | 0.40 |
Il13ra1 |
interleukin 13 receptor, alpha 1 |
243 |
0.94 |
| chr5_147747988_147748815 | 0.40 |
Gm43156 |
predicted gene 43156 |
2885 |
0.27 |
| chr11_119156502_119158281 | 0.40 |
Mir6934 |
microRNA 6934 |
3254 |
0.17 |
| chr7_29224845_29225704 | 0.39 |
Kcnk6 |
potassium inwardly-rectifying channel, subfamily K, member 6 |
6033 |
0.09 |
| chr15_79802268_79802553 | 0.39 |
Nptxr |
neuronal pentraxin receptor |
2299 |
0.17 |
| chr19_46320592_46320743 | 0.39 |
Psd |
pleckstrin and Sec7 domain containing |
2576 |
0.12 |
| chr3_11213105_11213577 | 0.39 |
Gm22547 |
predicted gene, 22547 |
59004 |
0.16 |
| chr16_42782105_42782459 | 0.39 |
4932412D23Rik |
RIKEN cDNA 4932412D23 gene |
93305 |
0.08 |
| chr7_74006482_74006835 | 0.39 |
St8sia2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
7016 |
0.2 |
| chr10_105689563_105689778 | 0.39 |
n-R5s80 |
nuclear encoded rRNA 5S 80 |
114527 |
0.05 |
| chr4_124576262_124576413 | 0.38 |
4933407E24Rik |
RIKEN cDNA 4933407E24 gene |
7147 |
0.19 |
| chr8_26268626_26269363 | 0.38 |
Gm31727 |
predicted gene, 31727 |
2017 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.3 | 1.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.3 | 1.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 0.9 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.3 | 0.8 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.2 | 1.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.3 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 0.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.4 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.1 | 0.5 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.2 | GO:0072197 | ureter morphogenesis(GO:0072197) |
| 0.1 | 0.4 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.3 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.4 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 1.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:1903802 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.1 | 0.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.2 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.1 | 0.2 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.6 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.9 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 4.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.3 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.1 | GO:0097104 | postsynaptic membrane assembly(GO:0097104) |
| 0.1 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0072079 | nephron tubule formation(GO:0072079) |
| 0.0 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.0 | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.7 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.0 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:0003096 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.0 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 1.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.0 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.0 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 1.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.2 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | Genes involved in Cytokine Signaling in Immune system |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |