Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
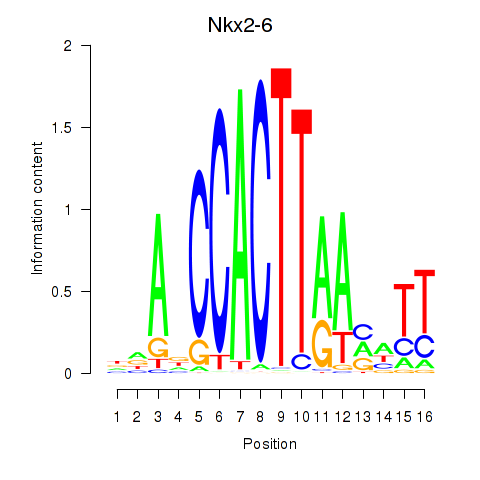
Results for Nkx2-6
Z-value: 0.67
Transcription factors associated with Nkx2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-6
|
ENSMUSG00000044186.9 | NK2 homeobox 6 |
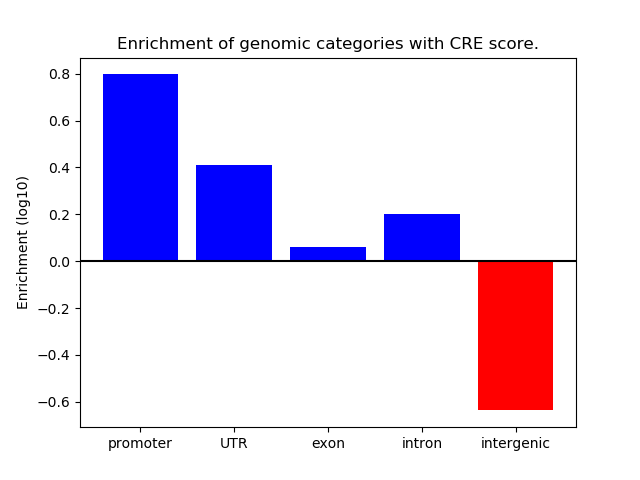
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_69169089_69169240 | Nkx2-6 | 2638 | 0.170934 | 0.25 | 5.4e-02 | Click! |
| chr14_69169675_69169826 | Nkx2-6 | 2052 | 0.197545 | -0.20 | 1.3e-01 | Click! |
| chr14_69171390_69172088 | Nkx2-6 | 63 | 0.566269 | -0.06 | 6.5e-01 | Click! |
Activity of the Nkx2-6 motif across conditions
Conditions sorted by the z-value of the Nkx2-6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
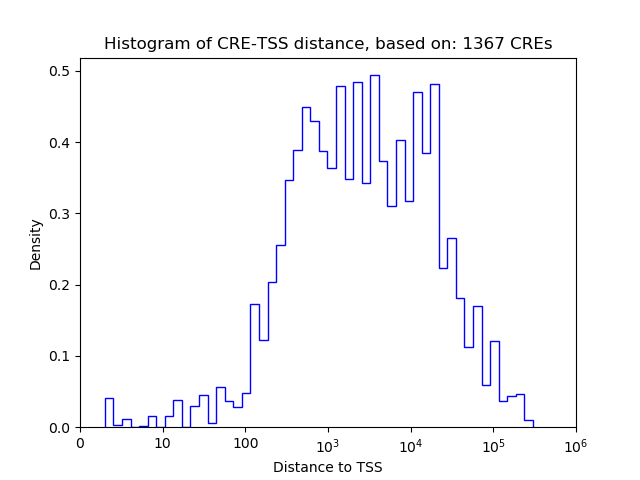
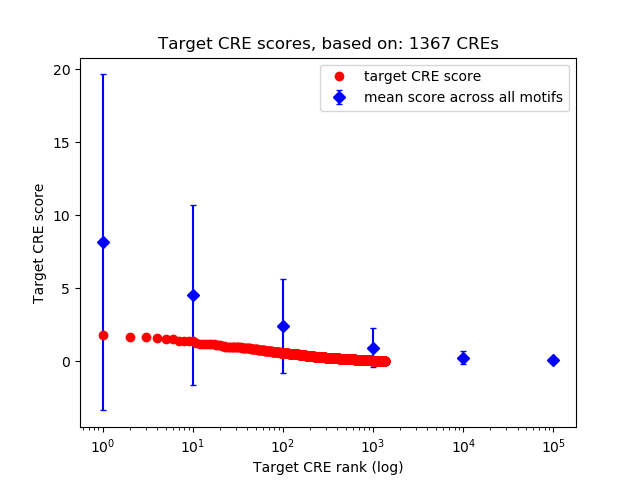
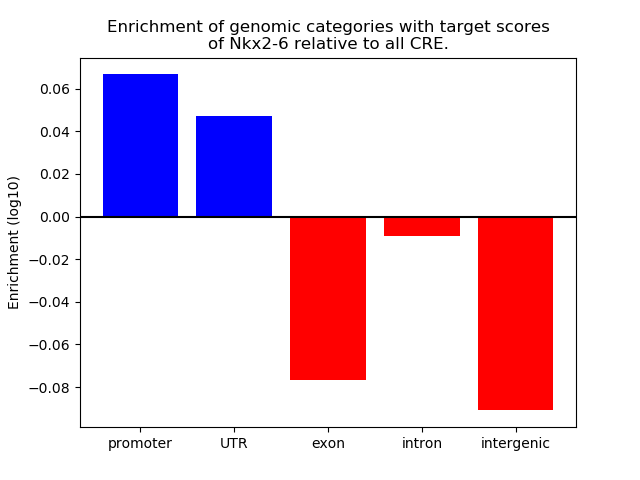
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_25941347_25942080 | 1.79 |
Retreg1 |
reticulophagy regulator 1 |
826 |
0.63 |
| chr10_108434849_108435510 | 1.69 |
Gm36283 |
predicted gene, 36283 |
1486 |
0.41 |
| chr8_28593556_28594184 | 1.68 |
Gm26795 |
predicted gene, 26795 |
338 |
0.92 |
| chr11_22170917_22171472 | 1.58 |
Ehbp1 |
EH domain binding protein 1 |
216 |
0.96 |
| chr1_40805583_40806390 | 1.54 |
Tmem182 |
transmembrane protein 182 |
385 |
0.84 |
| chr11_5802324_5803834 | 1.51 |
Pgam2 |
phosphoglycerate mutase 2 |
654 |
0.56 |
| chr3_59028229_59029161 | 1.42 |
Med12l |
mediator complex subunit 12-like |
21717 |
0.13 |
| chr9_111055888_111057545 | 1.41 |
Ccrl2 |
chemokine (C-C motif) receptor-like 2 |
530 |
0.62 |
| chr13_46422196_46422985 | 1.40 |
Rbm24 |
RNA binding motif protein 24 |
768 |
0.72 |
| chr10_122987387_122988205 | 1.36 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
1241 |
0.4 |
| chr17_69395929_69396464 | 1.24 |
Gm49894 |
predicted gene, 49894 |
11988 |
0.14 |
| chr19_34253411_34255499 | 1.21 |
Acta2 |
actin, alpha 2, smooth muscle, aorta |
225 |
0.92 |
| chr3_51708498_51709183 | 1.21 |
Gm10729 |
predicted gene 10729 |
15069 |
0.13 |
| chrX_7670339_7671484 | 1.18 |
Plp2 |
proteolipid protein 2 |
402 |
0.64 |
| chr6_142701068_142701298 | 1.18 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
1035 |
0.58 |
| chr15_80573378_80573655 | 1.16 |
Grap2 |
GRB2-related adaptor protein 2 |
922 |
0.53 |
| chr13_52584896_52585203 | 1.16 |
Syk |
spleen tyrosine kinase |
1546 |
0.51 |
| chr11_35006795_35007598 | 1.12 |
Gm25799 |
predicted gene, 25799 |
108179 |
0.06 |
| chr10_25432250_25432950 | 1.12 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
1 |
0.98 |
| chr10_13502765_13503111 | 1.08 |
Fuca2 |
fucosidase, alpha-L- 2, plasma |
1854 |
0.34 |
| chr3_98410919_98411390 | 1.08 |
Zfp697 |
zinc finger protein 697 |
28606 |
0.12 |
| chr8_111502069_111502570 | 1.02 |
Wdr59 |
WD repeat domain 59 |
11566 |
0.17 |
| chr3_60409216_60409437 | 1.01 |
Mbnl1 |
muscleblind like splicing factor 1 |
63504 |
0.12 |
| chr3_94933041_94933856 | 1.01 |
Selenbp1 |
selenium binding protein 1 |
289 |
0.81 |
| chr19_36732841_36733325 | 1.01 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
3570 |
0.26 |
| chr10_56468695_56469687 | 1.00 |
Gm9118 |
predicted gene 9118 |
28150 |
0.16 |
| chr3_144617679_144618017 | 1.00 |
Rpl9-ps8 |
ribosomal protein L9, pseudogene 8 |
8116 |
0.15 |
| chr17_72937768_72938085 | 1.00 |
Lbh |
limb-bud and heart |
16738 |
0.22 |
| chr1_55888894_55889264 | 1.00 |
9130227L01Rik |
RIKEN cDNA 9130227L01 gene |
42162 |
0.18 |
| chr3_103139237_103140483 | 0.99 |
Dennd2c |
DENN/MADD domain containing 2C |
466 |
0.74 |
| chr6_113748326_113749056 | 0.98 |
Sec13 |
SEC13 homolog, nuclear pore and COPII coat complex component |
7948 |
0.1 |
| chr9_77637137_77637792 | 0.98 |
Klhl31 |
kelch-like 31 |
964 |
0.54 |
| chr6_17761675_17761826 | 0.96 |
St7 |
suppression of tumorigenicity 7 |
12358 |
0.15 |
| chr2_69137426_69137980 | 0.94 |
Nostrin |
nitric oxide synthase trafficker |
1903 |
0.36 |
| chr2_68896037_68896304 | 0.93 |
Gm37159 |
predicted gene, 37159 |
4804 |
0.17 |
| chr3_57293411_57293693 | 0.93 |
Tm4sf1 |
transmembrane 4 superfamily member 1 |
151 |
0.96 |
| chr3_97075891_97076211 | 0.93 |
4930573H18Rik |
RIKEN cDNA 4930573H18 gene |
16734 |
0.15 |
| chr9_24642837_24642988 | 0.91 |
Cypt4 |
cysteine-rich perinuclear theca 4 |
17716 |
0.21 |
| chr8_89879797_89879982 | 0.91 |
Gm24212 |
predicted gene, 24212 |
212585 |
0.02 |
| chr2_91120400_91120551 | 0.90 |
Mybpc3 |
myosin binding protein C, cardiac |
2331 |
0.18 |
| chr4_117928946_117930077 | 0.89 |
Artn |
artemin |
14 |
0.96 |
| chr9_69458888_69459208 | 0.88 |
Mir3109 |
microRNA 3109 |
2104 |
0.2 |
| chr15_76666348_76670076 | 0.88 |
Foxh1 |
forkhead box H1 |
1590 |
0.15 |
| chr5_100214650_100214858 | 0.88 |
2310034O05Rik |
RIKEN cDNA 2310034O05 gene |
4046 |
0.22 |
| chr10_79824203_79824406 | 0.87 |
Misp |
mitotic spindle positioning |
383 |
0.64 |
| chr6_112459682_112460792 | 0.87 |
Cav3 |
caveolin 3 |
732 |
0.65 |
| chr13_41019834_41020226 | 0.87 |
Tmem14c |
transmembrane protein 14C |
3738 |
0.15 |
| chr11_12308516_12308667 | 0.86 |
Gm12002 |
predicted gene 12002 |
5923 |
0.29 |
| chr19_43916774_43917584 | 0.84 |
Gm50217 |
predicted gene, 50217 |
3294 |
0.17 |
| chrX_141476575_141476992 | 0.84 |
Col4a5 |
collagen, type IV, alpha 5 |
1373 |
0.41 |
| chr17_75435976_75437284 | 0.84 |
Rasgrp3 |
RAS, guanyl releasing protein 3 |
704 |
0.77 |
| chr1_150984135_150984286 | 0.83 |
Hmcn1 |
hemicentin 1 |
8841 |
0.19 |
| chr5_114146628_114147196 | 0.80 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
377 |
0.78 |
| chr4_32890178_32890640 | 0.80 |
Gm11941 |
predicted gene 11941 |
2277 |
0.29 |
| chr19_6384347_6385874 | 0.79 |
Pygm |
muscle glycogen phosphorylase |
695 |
0.45 |
| chr1_133492302_133493599 | 0.79 |
Gm8596 |
predicted gene 8596 |
36529 |
0.14 |
| chr1_52251127_52251278 | 0.79 |
Gls |
glutaminase |
17970 |
0.19 |
| chr15_75111128_75111422 | 0.78 |
Ly6c2 |
lymphocyte antigen 6 complex, locus C2 |
422 |
0.74 |
| chr10_69571124_69572441 | 0.78 |
Gm46231 |
predicted gene, 46231 |
5421 |
0.27 |
| chr5_77126646_77126869 | 0.77 |
Gm15831 |
predicted gene 15831 |
4409 |
0.15 |
| chr13_5897408_5897746 | 0.76 |
Gm47507 |
predicted gene, 47507 |
10468 |
0.16 |
| chr6_31114239_31115170 | 0.74 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
3784 |
0.15 |
| chr1_74154525_74154676 | 0.73 |
Cxcr2 |
chemokine (C-X-C motif) receptor 2 |
609 |
0.64 |
| chr10_95970803_95971600 | 0.73 |
Eea1 |
early endosome antigen 1 |
30444 |
0.15 |
| chr14_79536799_79537268 | 0.71 |
Elf1 |
E74-like factor 1 |
21335 |
0.14 |
| chr8_57320946_57324000 | 0.71 |
Hand2os1 |
Hand2, opposite strand 1 |
1245 |
0.3 |
| chr5_117133717_117134877 | 0.70 |
Taok3 |
TAO kinase 3 |
656 |
0.62 |
| chr10_63366309_63366946 | 0.70 |
Gm16145 |
predicted gene 16145 |
12774 |
0.12 |
| chr6_51171741_51172819 | 0.69 |
Mir148a |
microRNA 148a |
97630 |
0.07 |
| chr15_12323754_12324089 | 0.69 |
1810049J17Rik |
RIKEN cDNA 1810049J17 gene |
2022 |
0.26 |
| chr19_25383229_25384087 | 0.69 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
11645 |
0.21 |
| chr2_119601570_119601760 | 0.68 |
Oip5os1 |
Opa interacting protein 5, opposite strand 1 |
7362 |
0.11 |
| chr5_149068416_149068775 | 0.68 |
Hmgb1 |
high mobility group box 1 |
15082 |
0.11 |
| chr10_42571122_42571273 | 0.68 |
Nr2e1 |
nuclear receptor subfamily 2, group E, member 1 |
2369 |
0.3 |
| chr16_36157136_36157365 | 0.68 |
Stfa2l1 |
stefin A2 like 1 |
439 |
0.63 |
| chr10_18886828_18886979 | 0.67 |
Gm48483 |
predicted gene, 48483 |
126 |
0.97 |
| chr13_45731554_45731705 | 0.66 |
Gm47460 |
predicted gene, 47460 |
17943 |
0.26 |
| chr12_51709478_51709629 | 0.65 |
Strn3 |
striatin, calmodulin binding protein 3 |
17656 |
0.16 |
| chr5_67777924_67778180 | 0.65 |
Atp8a1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
1293 |
0.4 |
| chr11_32280517_32281064 | 0.64 |
Hba-a1 |
hemoglobin alpha, adult chain 1 |
2721 |
0.14 |
| chr5_29030494_29030763 | 0.64 |
Rnf32 |
ring finger protein 32 |
165364 |
0.03 |
| chr9_113970028_113970753 | 0.64 |
Gm47950 |
predicted gene, 47950 |
608 |
0.61 |
| chr12_105034229_105034527 | 0.64 |
Glrx5 |
glutaredoxin 5 |
838 |
0.39 |
| chr9_53850550_53850766 | 0.63 |
Sln |
sarcolipin |
494 |
0.79 |
| chr18_10785293_10785854 | 0.63 |
Mir1a-2 |
microRNA 1a-2 |
21 |
0.55 |
| chr8_70803797_70804348 | 0.63 |
Mast3 |
microtubule associated serine/threonine kinase 3 |
910 |
0.3 |
| chrX_107149448_107150320 | 0.61 |
P2ry10b |
purinergic receptor P2Y, G-protein coupled 10B |
269 |
0.92 |
| chrX_136144132_136144311 | 0.61 |
Bex4 |
brain expressed X-linked 4 |
5225 |
0.13 |
| chr5_99339481_99340596 | 0.61 |
Gm35394 |
predicted gene, 35394 |
65943 |
0.12 |
| chr8_26329826_26330767 | 0.61 |
Gm31784 |
predicted gene, 31784 |
17962 |
0.15 |
| chr16_30520227_30520435 | 0.61 |
Tmem44 |
transmembrane protein 44 |
20578 |
0.18 |
| chr18_74502159_74502310 | 0.60 |
1700120E14Rik |
RIKEN cDNA 1700120E14 gene |
28872 |
0.18 |
| chr8_74871836_74872493 | 0.60 |
Isx |
intestine specific homeobox |
1009 |
0.61 |
| chr9_40877257_40878943 | 0.60 |
Bsx |
brain specific homeobox |
3973 |
0.16 |
| chr18_65083730_65084940 | 0.60 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
1281 |
0.54 |
| chr13_107065067_107065474 | 0.59 |
Gm31452 |
predicted gene, 31452 |
1575 |
0.35 |
| chr4_31404597_31405733 | 0.59 |
Gm11922 |
predicted gene 11922 |
24693 |
0.28 |
| chr4_45965494_45966277 | 0.59 |
Tdrd7 |
tudor domain containing 7 |
551 |
0.79 |
| chr11_49247391_49247809 | 0.59 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
117 |
0.94 |
| chr4_115059803_115061295 | 0.57 |
Tal1 |
T cell acute lymphocytic leukemia 1 |
1041 |
0.47 |
| chr17_81740807_81741112 | 0.57 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
2582 |
0.38 |
| chrX_140540281_140540484 | 0.56 |
Tsc22d3 |
TSC22 domain family, member 3 |
2286 |
0.31 |
| chr12_77483355_77483606 | 0.56 |
Gm48177 |
predicted gene, 48177 |
42918 |
0.13 |
| chr11_65265838_65266454 | 0.56 |
Myocd |
myocardin |
3708 |
0.26 |
| chr16_23028396_23028918 | 0.56 |
Kng2 |
kininogen 2 |
352 |
0.74 |
| chr5_51923307_51923594 | 0.56 |
Gm43175 |
predicted gene 43175 |
12519 |
0.18 |
| chr6_122609501_122610041 | 0.55 |
Gdf3 |
growth differentiation factor 3 |
53 |
0.95 |
| chr8_68119212_68120289 | 0.55 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
1563 |
0.51 |
| chr1_164048817_164048989 | 0.55 |
Sele |
selectin, endothelial cell |
669 |
0.62 |
| chr8_79641995_79642830 | 0.55 |
Otud4 |
OTU domain containing 4 |
2708 |
0.24 |
| chr19_55252255_55252406 | 0.55 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
370 |
0.85 |
| chr8_94379515_94379666 | 0.55 |
Gm15889 |
predicted gene 15889 |
1669 |
0.22 |
| chr8_53637135_53637740 | 0.54 |
Neil3 |
nei like 3 (E. coli) |
1241 |
0.61 |
| chr13_12320716_12320867 | 0.54 |
Actn2 |
actinin alpha 2 |
19933 |
0.16 |
| chr12_33061431_33061850 | 0.53 |
Cdhr3 |
cadherin-related family member 3 |
152 |
0.89 |
| chr10_7472533_7472784 | 0.53 |
Ulbp1 |
UL16 binding protein 1 |
683 |
0.71 |
| chr1_165462988_165463362 | 0.53 |
Mpc2 |
mitochondrial pyruvate carrier 2 |
1810 |
0.18 |
| chr6_53986001_53986442 | 0.53 |
4921529L05Rik |
RIKEN cDNA 4921529L05 gene |
7492 |
0.2 |
| chr1_77213373_77213909 | 0.53 |
Gm38265 |
predicted gene, 38265 |
66649 |
0.12 |
| chr2_145822082_145822862 | 0.52 |
Rin2 |
Ras and Rab interactor 2 |
183 |
0.96 |
| chr1_50962507_50962885 | 0.52 |
Tmeff2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
35177 |
0.17 |
| chr14_34273565_34273716 | 0.52 |
Shld2 |
shieldin complex subunit 2 |
619 |
0.54 |
| chrX_169688414_169688708 | 0.52 |
Mid1 |
midline 1 |
3362 |
0.28 |
| chr8_45681436_45681587 | 0.52 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
6822 |
0.23 |
| chr17_88627411_88628033 | 0.52 |
Ston1 |
stonin 1 |
358 |
0.85 |
| chr7_24851355_24851650 | 0.51 |
Gm18207 |
predicted gene, 18207 |
9444 |
0.09 |
| chr9_75775275_75776608 | 0.51 |
Bmp5 |
bone morphogenetic protein 5 |
577 |
0.78 |
| chr1_133799248_133799949 | 0.51 |
Atp2b4 |
ATPase, Ca++ transporting, plasma membrane 4 |
1438 |
0.33 |
| chr6_48707462_48707684 | 0.51 |
Gimap6 |
GTPase, IMAP family member 6 |
623 |
0.46 |
| chr1_131638462_131638811 | 0.51 |
Ctse |
cathepsin E |
142 |
0.95 |
| chr1_167269338_167269683 | 0.51 |
Uck2 |
uridine-cytidine kinase 2 |
15091 |
0.11 |
| chr2_34944829_34945211 | 0.50 |
Traf1 |
TNF receptor-associated factor 1 |
13116 |
0.11 |
| chr4_45489091_45489458 | 0.50 |
Shb |
src homology 2 domain-containing transforming protein B |
129 |
0.96 |
| chr7_45233983_45235751 | 0.50 |
Cd37 |
CD37 antigen |
591 |
0.44 |
| chr6_87495313_87495551 | 0.49 |
Arhgap25 |
Rho GTPase activating protein 25 |
882 |
0.56 |
| chr10_76621571_76623890 | 0.49 |
Col6a2 |
collagen, type VI, alpha 2 |
595 |
0.68 |
| chr7_133700764_133701966 | 0.49 |
Uros |
uroporphyrinogen III synthase |
1173 |
0.35 |
| chr17_70997929_70998765 | 0.48 |
Myl12a |
myosin, light chain 12A, regulatory, non-sarcomeric |
216 |
0.88 |
| chr13_110882860_110883156 | 0.48 |
Gm38397 |
predicted gene, 38397 |
20491 |
0.14 |
| chr14_54711789_54711940 | 0.48 |
Cebpe |
CCAAT/enhancer binding protein (C/EBP), epsilon |
310 |
0.79 |
| chr10_118858762_118859192 | 0.48 |
4932442E05Rik |
RIKEN cDNA 4932442E05 gene |
1849 |
0.25 |
| chr11_102145120_102148094 | 0.48 |
Nags |
N-acetylglutamate synthase |
241 |
0.58 |
| chr7_112268841_112268992 | 0.47 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
2383 |
0.41 |
| chr6_119194389_119194967 | 0.47 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1553 |
0.37 |
| chr6_125349409_125350825 | 0.47 |
Tnfrsf1a |
tumor necrosis factor receptor superfamily, member 1a |
394 |
0.77 |
| chr10_79988072_79989720 | 0.47 |
Cnn2 |
calponin 2 |
273 |
0.76 |
| chr3_101395066_101395306 | 0.47 |
Gm42939 |
predicted gene 42939 |
12607 |
0.14 |
| chr2_69672410_69673170 | 0.47 |
Klhl41 |
kelch-like 41 |
2670 |
0.21 |
| chr7_98341637_98342055 | 0.46 |
Tsku |
tsukushi, small leucine rich proteoglycan |
18233 |
0.16 |
| chr12_56695295_56695580 | 0.46 |
Pax9 |
paired box 9 |
202 |
0.92 |
| chr14_119285808_119285959 | 0.46 |
Gm6087 |
predicted gene 6087 |
7459 |
0.3 |
| chr10_68132791_68133177 | 0.46 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
3642 |
0.31 |
| chr6_33055424_33055891 | 0.46 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
4560 |
0.2 |
| chr12_51350200_51351160 | 0.46 |
G2e3 |
G2/M-phase specific E3 ubiquitin ligase |
2308 |
0.33 |
| chr3_51042270_51042900 | 0.46 |
Gm29230 |
predicted gene 29230 |
32271 |
0.16 |
| chr4_109664033_109664623 | 0.46 |
Cdkn2c |
cyclin dependent kinase inhibitor 2C |
1044 |
0.46 |
| chr11_109343750_109344455 | 0.45 |
1700096J18Rik |
RIKEN cDNA 1700096J18 gene |
2737 |
0.2 |
| chr5_119673827_119675890 | 0.45 |
Tbx3 |
T-box 3 |
587 |
0.67 |
| chr16_38709840_38710271 | 0.45 |
Arhgap31 |
Rho GTPase activating protein 31 |
2843 |
0.21 |
| chr2_97601844_97601995 | 0.44 |
Gm44320 |
predicted gene, 44320 |
25133 |
0.27 |
| chr6_144480846_144480997 | 0.44 |
Sox5os2 |
SRY (sex determining region Y)-box 5, opposite strand 2 |
304 |
0.94 |
| chr15_40673370_40673521 | 0.44 |
Zfpm2 |
zinc finger protein, multitype 2 |
8455 |
0.3 |
| chr8_40905205_40905856 | 0.44 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
7092 |
0.17 |
| chr4_120612414_120612969 | 0.44 |
Gm12860 |
predicted gene 12860 |
3149 |
0.19 |
| chr14_54228146_54228679 | 0.44 |
Traj1 |
T cell receptor alpha joining 1 |
9598 |
0.08 |
| chr17_47909108_47910390 | 0.43 |
Gm15556 |
predicted gene 15556 |
12629 |
0.13 |
| chrX_99045264_99045888 | 0.42 |
Stard8 |
START domain containing 8 |
1592 |
0.43 |
| chr5_17887492_17887771 | 0.42 |
Cd36 |
CD36 molecule |
1116 |
0.65 |
| chr1_91052629_91054398 | 0.42 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
6 |
0.98 |
| chr4_46065775_46065926 | 0.41 |
Tmod1 |
tropomodulin 1 |
11277 |
0.19 |
| chr6_142878886_142879637 | 0.41 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
3779 |
0.24 |
| chr16_55381050_55381215 | 0.41 |
Zpld1 |
zona pellucida like domain containing 1 |
83146 |
0.1 |
| chr10_60828323_60829684 | 0.41 |
Unc5b |
unc-5 netrin receptor B |
2379 |
0.28 |
| chr4_33379003_33379165 | 0.41 |
Rngtt |
RNA guanylyltransferase and 5'-phosphatase |
48070 |
0.12 |
| chr14_14067690_14068738 | 0.40 |
Atxn7 |
ataxin 7 |
19255 |
0.17 |
| chr3_53165376_53165642 | 0.40 |
Gm43803 |
predicted gene 43803 |
52098 |
0.09 |
| chr4_89446020_89446496 | 0.40 |
Gm12608 |
predicted gene 12608 |
1614 |
0.4 |
| chr18_39481253_39481404 | 0.40 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
5904 |
0.28 |
| chr7_128527029_128527321 | 0.40 |
Bag3 |
BCL2-associated athanogene 3 |
3559 |
0.15 |
| chr11_96318060_96319501 | 0.39 |
Hoxb4 |
homeobox B4 |
513 |
0.51 |
| chr19_45223479_45224523 | 0.39 |
Lbx1 |
ladybird homeobox 1 |
11811 |
0.18 |
| chr8_126833216_126834027 | 0.39 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
5612 |
0.23 |
| chr2_92486138_92486303 | 0.38 |
Gm13814 |
predicted gene 13814 |
2294 |
0.22 |
| chr17_26841784_26842428 | 0.38 |
Nkx2-5 |
NK2 homeobox 5 |
541 |
0.65 |
| chr2_83733967_83734585 | 0.38 |
Itgav |
integrin alpha V |
9721 |
0.15 |
| chr10_56384887_56385255 | 0.38 |
Gja1 |
gap junction protein, alpha 1 |
5983 |
0.21 |
| chr18_23748497_23748648 | 0.38 |
Gm15972 |
predicted gene 15972 |
3733 |
0.2 |
| chr5_43981645_43981861 | 0.38 |
Fgfbp1 |
fibroblast growth factor binding protein 1 |
11 |
0.96 |
| chr3_27690391_27690623 | 0.38 |
Fndc3b |
fibronectin type III domain containing 3B |
19891 |
0.25 |
| chr18_50055297_50055988 | 0.38 |
C030005K06Rik |
RIKEN cDNA C030005K06 gene |
1798 |
0.32 |
| chr16_32610201_32610589 | 0.38 |
Tfrc |
transferrin receptor |
1145 |
0.41 |
| chr19_25607525_25608179 | 0.37 |
Dmrt3 |
doublesex and mab-3 related transcription factor 3 |
2449 |
0.33 |
| chr10_42247495_42248433 | 0.37 |
Foxo3 |
forkhead box O3 |
10402 |
0.26 |
| chr1_178798383_178799679 | 0.37 |
Kif26b |
kinesin family member 26B |
593 |
0.81 |
| chr5_113648616_113649038 | 0.37 |
Cmklr1 |
chemokine-like receptor 1 |
1569 |
0.33 |
| chr7_139991869_139992020 | 0.37 |
Adam8 |
a disintegrin and metallopeptidase domain 8 |
564 |
0.55 |
| chr12_111421008_111421645 | 0.37 |
Exoc3l4 |
exocyst complex component 3-like 4 |
235 |
0.88 |
| chr2_120147858_120148056 | 0.36 |
Ehd4 |
EH-domain containing 4 |
6505 |
0.18 |
| chr5_20583581_20583870 | 0.36 |
Gm3544 |
predicted gene 3544 |
4871 |
0.26 |
| chr2_148438023_148438174 | 0.36 |
Cd93 |
CD93 antigen |
5465 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.3 | 0.8 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 0.6 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.2 | 0.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.5 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.1 | 0.6 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.5 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 0.4 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.1 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.1 | 0.3 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.5 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.1 | 0.6 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 | 0.2 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.3 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.5 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.5 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.1 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.1 | 0.1 | GO:0003195 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.3 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.1 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.3 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.2 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.2 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 1.2 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.1 | 0.6 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.4 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.6 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.4 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0002836 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.0 | 0.4 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.5 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0019661 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.4 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.8 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.0 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.5 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 1.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 0.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.4 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.3 | GO:0018502 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.1 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0044682 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0019113 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.4 | GO:0015928 | fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |