Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
Results for Nr0b1
Z-value: 0.51
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.4 | nuclear receptor subfamily 0, group B, member 1 |
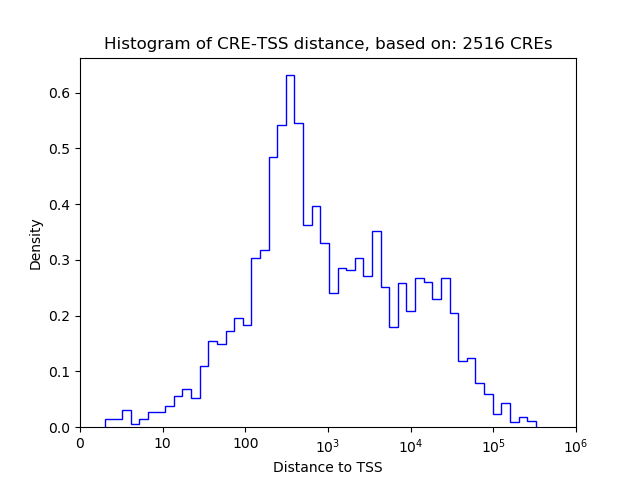
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_86191680_86192868 | Nr0b1 | 510 | 0.816189 | 0.25 | 5.5e-02 | Click! |
Activity of the Nr0b1 motif across conditions
Conditions sorted by the z-value of the Nr0b1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
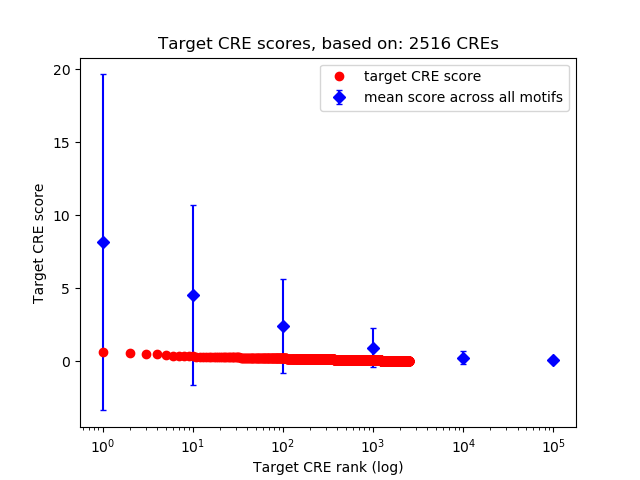
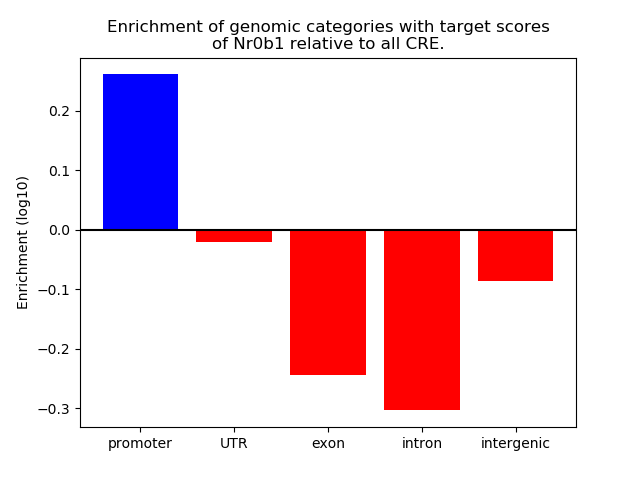
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_119803355_119804193 | 0.63 |
1700021F13Rik |
RIKEN cDNA 1700021F13 gene |
3893 |
0.21 |
| chr2_172936311_172937024 | 0.56 |
Bmp7 |
bone morphogenetic protein 7 |
3425 |
0.23 |
| chr17_32165675_32166119 | 0.51 |
Notch3 |
notch 3 |
955 |
0.45 |
| chr3_9347262_9347787 | 0.48 |
C030034L19Rik |
RIKEN cDNA C030034L19 gene |
55540 |
0.13 |
| chr6_63256981_63257723 | 0.46 |
9330118I20Rik |
RIKEN cDNA 9330118I20 gene |
273 |
0.71 |
| chr11_5802324_5803834 | 0.37 |
Pgam2 |
phosphoglycerate mutase 2 |
654 |
0.56 |
| chr18_76544225_76544908 | 0.36 |
Gm31933 |
predicted gene, 31933 |
88304 |
0.1 |
| chr4_134370897_134371720 | 0.34 |
Extl1 |
exostosin-like glycosyltransferase 1 |
1250 |
0.33 |
| chr14_54986330_54986529 | 0.33 |
Myh7 |
myosin, heavy polypeptide 7, cardiac muscle, beta |
989 |
0.26 |
| chr10_78787171_78788340 | 0.33 |
Slc1a6 |
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
7053 |
0.14 |
| chr16_94721948_94722518 | 0.33 |
Gm41505 |
predicted gene, 41505 |
339 |
0.89 |
| chr17_26840808_26841709 | 0.32 |
Nkx2-5 |
NK2 homeobox 5 |
307 |
0.83 |
| chr6_29434692_29436459 | 0.31 |
Flnc |
filamin C, gamma |
2299 |
0.16 |
| chr9_53402629_53402979 | 0.30 |
4930550C14Rik |
RIKEN cDNA 4930550C14 gene |
479 |
0.73 |
| chr2_167239298_167240893 | 0.30 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
494 |
0.75 |
| chr3_138066011_138066302 | 0.30 |
1110002E22Rik |
RIKEN cDNA 1110002E22 gene |
1104 |
0.33 |
| chr15_54278043_54279091 | 0.29 |
Tnfrsf11b |
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
83 |
0.98 |
| chr14_55826099_55826996 | 0.29 |
Nfatc4 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
491 |
0.63 |
| chr9_21851806_21852608 | 0.29 |
Dock6 |
dedicator of cytokinesis 6 |
409 |
0.74 |
| chr8_57324050_57324269 | 0.29 |
Hand2os1 |
Hand2, opposite strand 1 |
74 |
0.96 |
| chr10_109921017_109921811 | 0.28 |
Nav3 |
neuron navigator 3 |
17391 |
0.27 |
| chr6_54329128_54329297 | 0.28 |
9130019P16Rik |
RIKEN cDNA 9130019P16 gene |
1871 |
0.29 |
| chr16_4522019_4522667 | 0.28 |
Srl |
sarcalumenin |
720 |
0.62 |
| chr16_4641802_4643186 | 0.28 |
Dnaja3 |
DnaJ heat shock protein family (Hsp40) member A3 |
2505 |
0.16 |
| chr10_22813360_22814229 | 0.28 |
Gm10824 |
predicted gene 10824 |
1955 |
0.28 |
| chr13_93612831_93613054 | 0.28 |
Gm15622 |
predicted gene 15622 |
12440 |
0.16 |
| chr13_46419789_46420118 | 0.27 |
Rbm24 |
RNA binding motif protein 24 |
139 |
0.97 |
| chr7_112225854_112227329 | 0.27 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
701 |
0.78 |
| chr3_85186481_85187125 | 0.27 |
Gm38313 |
predicted gene, 38313 |
9644 |
0.24 |
| chr2_30995683_30996883 | 0.27 |
Usp20 |
ubiquitin specific peptidase 20 |
235 |
0.9 |
| chr15_8279541_8279705 | 0.26 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
32520 |
0.18 |
| chr6_82772676_82772834 | 0.26 |
Hk2 |
hexokinase 2 |
233 |
0.9 |
| chr1_162219410_162219749 | 0.26 |
Dnm3os |
dynamin 3, opposite strand |
289 |
0.86 |
| chr3_34795242_34796350 | 0.26 |
Gm38509 |
predicted gene, 38509 |
23721 |
0.17 |
| chr7_25787527_25787868 | 0.26 |
Axl |
AXL receptor tyrosine kinase |
125 |
0.92 |
| chr9_95256675_95257394 | 0.25 |
Slc9a9 |
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
135348 |
0.04 |
| chr4_126627040_126627629 | 0.25 |
5730409E04Rik |
RIKEN cDNA 5730409E04Rik gene |
17480 |
0.12 |
| chr11_102606785_102607497 | 0.25 |
Fzd2 |
frizzled class receptor 2 |
2745 |
0.14 |
| chr1_134036991_134037429 | 0.25 |
Fmod |
fibromodulin |
44 |
0.97 |
| chr3_138069501_138070764 | 0.25 |
1110002E22Rik |
RIKEN cDNA 1110002E22 gene |
1465 |
0.3 |
| chr9_77345728_77346390 | 0.25 |
Mlip |
muscular LMNA-interacting protein |
1734 |
0.32 |
| chr1_69107020_69107405 | 0.25 |
Gm16076 |
predicted gene 16076 |
472 |
0.52 |
| chr5_19228089_19228240 | 0.25 |
Magi2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
767 |
0.56 |
| chr3_82144651_82145810 | 0.25 |
Gucy1a1 |
guanylate cyclase 1, soluble, alpha 1 |
157 |
0.96 |
| chr8_48843313_48843722 | 0.24 |
Gm19744 |
predicted gene, 19744 |
148 |
0.81 |
| chr6_53288842_53289356 | 0.24 |
Creb5 |
cAMP responsive element binding protein 5 |
1324 |
0.55 |
| chr2_85049344_85049658 | 0.24 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
959 |
0.42 |
| chr2_110361610_110361920 | 0.24 |
Fibin |
fin bud initiation factor homolog (zebrafish) |
1418 |
0.47 |
| chr13_38153771_38154277 | 0.23 |
Gm10129 |
predicted gene 10129 |
2232 |
0.25 |
| chr17_11912724_11913972 | 0.23 |
Prkn |
parkin RBR E3 ubiquitin protein ligase |
68138 |
0.13 |
| chr3_84189998_84191469 | 0.23 |
Trim2 |
tripartite motif-containing 2 |
210 |
0.94 |
| chr19_11818257_11818813 | 0.23 |
Stx3 |
syntaxin 3 |
186 |
0.9 |
| chr2_79646122_79646273 | 0.23 |
Itprid2 |
ITPR interacting domain containing 2 |
7015 |
0.28 |
| chr19_4439491_4439665 | 0.23 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
19 |
0.64 |
| chr8_81856918_81857264 | 0.23 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
723 |
0.73 |
| chr7_107665595_107666983 | 0.22 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
978 |
0.52 |
| chrX_12116163_12116355 | 0.22 |
Bcor |
BCL6 interacting corepressor |
12091 |
0.26 |
| chr8_102780577_102781067 | 0.22 |
Gm45258 |
predicted gene 45258 |
220 |
0.93 |
| chr13_103355936_103356429 | 0.22 |
Gm5454 |
predicted gene 5454 |
70 |
0.97 |
| chr2_31313954_31314959 | 0.22 |
Hmcn2 |
hemicentin 2 |
41 |
0.98 |
| chr5_18360537_18360783 | 0.22 |
Gnai1 |
guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
305 |
0.93 |
| chr5_132541084_132542037 | 0.22 |
Auts2 |
autism susceptibility candidate 2 |
263 |
0.87 |
| chr8_22507762_22508195 | 0.22 |
Slc20a2 |
solute carrier family 20, member 2 |
1752 |
0.29 |
| chr14_46389448_46390599 | 0.22 |
Bmp4 |
bone morphogenetic protein 4 |
512 |
0.7 |
| chr19_53808045_53808428 | 0.22 |
Rbm20 |
RNA binding motif protein 20 |
14928 |
0.16 |
| chr17_42975571_42975917 | 0.22 |
Tnfrsf21 |
tumor necrosis factor receptor superfamily, member 21 |
40811 |
0.19 |
| chr6_138171729_138171880 | 0.22 |
Mgst1 |
microsomal glutathione S-transferase 1 |
28950 |
0.23 |
| chr13_74362852_74363978 | 0.22 |
Lrrc14b |
leucine rich repeat containing 14B |
590 |
0.54 |
| chr6_4506970_4507670 | 0.22 |
Gm43921 |
predicted gene, 43921 |
1233 |
0.34 |
| chr11_116885438_116886127 | 0.22 |
Mfsd11 |
major facilitator superfamily domain containing 11 |
17414 |
0.12 |
| chr7_142472156_142472970 | 0.22 |
Lsp1 |
lymphocyte specific 1 |
412 |
0.73 |
| chr2_91120847_91121416 | 0.22 |
Mybpc3 |
myosin binding protein C, cardiac |
2987 |
0.16 |
| chr12_108837523_108838185 | 0.21 |
Slc25a47 |
solute carrier family 25, member 47 |
790 |
0.33 |
| chr15_89557488_89558668 | 0.21 |
Shank3 |
SH3 and multiple ankyrin repeat domains 3 |
1376 |
0.31 |
| chr5_13280340_13280537 | 0.21 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
116346 |
0.06 |
| chr2_30902329_30903582 | 0.21 |
Ptges |
prostaglandin E synthase |
342 |
0.83 |
| chr2_163285470_163285918 | 0.21 |
Tox2 |
TOX high mobility group box family member 2 |
34684 |
0.16 |
| chr1_134513098_134514080 | 0.21 |
Gm15454 |
predicted gene 15454 |
4492 |
0.12 |
| chr10_111419452_111420510 | 0.21 |
Nap1l1 |
nucleosome assembly protein 1-like 1 |
53242 |
0.1 |
| chr17_66232919_66233799 | 0.21 |
Gm49932 |
predicted gene, 49932 |
1586 |
0.29 |
| chr18_58210827_58211095 | 0.21 |
Fbn2 |
fibrillin 2 |
474 |
0.86 |
| chr16_14705845_14706609 | 0.21 |
Snai2 |
snail family zinc finger 2 |
375 |
0.9 |
| chr16_35659651_35660734 | 0.21 |
Sema5b |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
757 |
0.67 |
| chr17_68002299_68002969 | 0.21 |
Arhgap28 |
Rho GTPase activating protein 28 |
1486 |
0.54 |
| chr2_4298620_4299406 | 0.21 |
Frmd4a |
FERM domain containing 4A |
1505 |
0.32 |
| chr8_120231782_120232001 | 0.21 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
3435 |
0.2 |
| chr5_125139990_125141281 | 0.21 |
Ncor2 |
nuclear receptor co-repressor 2 |
15735 |
0.2 |
| chr2_180332745_180333375 | 0.21 |
Gata5os |
GATA binding protein 5, opposite strand |
203 |
0.89 |
| chr5_43515484_43516406 | 0.20 |
C1qtnf7 |
C1q and tumor necrosis factor related protein 7 |
183 |
0.94 |
| chr9_30921334_30922737 | 0.20 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
417 |
0.86 |
| chr5_24601266_24602082 | 0.20 |
Smarcd3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
226 |
0.86 |
| chr5_129854658_129854926 | 0.20 |
Sumf2 |
sulfatase modifying factor 2 |
2281 |
0.16 |
| chr9_56712934_56713139 | 0.20 |
Lingo1 |
leucine rich repeat and Ig domain containing 1 |
27783 |
0.18 |
| chr10_103367601_103367825 | 0.20 |
Slc6a15 |
solute carrier family 6 (neurotransmitter transporter), member 15 |
70 |
0.98 |
| chr14_51740214_51741404 | 0.20 |
Gm49261 |
predicted gene, 49261 |
296 |
0.81 |
| chr3_128990459_128991248 | 0.20 |
Gm9387 |
predicted pseudogene 9387 |
35755 |
0.19 |
| chr5_66746743_66746894 | 0.20 |
Limch1 |
LIM and calponin homology domains 1 |
929 |
0.55 |
| chr6_88200114_88201718 | 0.20 |
Gata2 |
GATA binding protein 2 |
3 |
0.96 |
| chr7_24499513_24500150 | 0.20 |
Cadm4 |
cell adhesion molecule 4 |
130 |
0.91 |
| chr19_34253411_34255499 | 0.20 |
Acta2 |
actin, alpha 2, smooth muscle, aorta |
225 |
0.92 |
| chr13_32966469_32966620 | 0.20 |
Serpinb6b |
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
1041 |
0.43 |
| chr2_30469737_30470841 | 0.20 |
Ier5l |
immediate early response 5-like |
3930 |
0.16 |
| chr4_155839735_155840422 | 0.20 |
Mxra8os |
matrix-remodelling associated 8, opposite strand |
100 |
0.67 |
| chr2_67565793_67566876 | 0.19 |
B3galt1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
463 |
0.84 |
| chr10_22158248_22159850 | 0.19 |
H60b |
histocompatibility 60b |
463 |
0.42 |
| chr7_30291325_30291668 | 0.19 |
Clip3 |
CAP-GLY domain containing linker protein 3 |
232 |
0.8 |
| chr7_112180336_112180681 | 0.19 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
21451 |
0.22 |
| chr10_67939481_67939695 | 0.19 |
Zfp365 |
zinc finger protein 365 |
26926 |
0.16 |
| chr14_105893244_105894185 | 0.19 |
Spry2 |
sprouty RTK signaling antagonist 2 |
3105 |
0.29 |
| chr16_30540363_30541394 | 0.19 |
Tmem44 |
transmembrane protein 44 |
31 |
0.98 |
| chr11_105293049_105293533 | 0.19 |
Mrc2 |
mannose receptor, C type 2 |
623 |
0.74 |
| chr4_31823845_31825033 | 0.19 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
139658 |
0.05 |
| chr4_34436178_34436537 | 0.19 |
Gm12351 |
predicted gene 12351 |
21533 |
0.16 |
| chr1_74090075_74091356 | 0.19 |
Tns1 |
tensin 1 |
636 |
0.71 |
| chr7_18957439_18958650 | 0.19 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
29356 |
0.06 |
| chr15_76194294_76195998 | 0.19 |
Plec |
plectin |
564 |
0.56 |
| chr4_42950234_42951162 | 0.19 |
Dnajb5 |
DnaJ heat shock protein family (Hsp40) member B5 |
258 |
0.87 |
| chr6_128285700_128286493 | 0.19 |
Gm21893 |
predicted gene, 21893 |
8006 |
0.09 |
| chr1_134801190_134801791 | 0.18 |
Gm37949 |
predicted gene, 37949 |
495 |
0.7 |
| chr1_135793443_135794350 | 0.18 |
Tnni1 |
troponin I, skeletal, slow 1 |
5506 |
0.15 |
| chr4_11705100_11705674 | 0.18 |
Gem |
GTP binding protein (gene overexpressed in skeletal muscle) |
420 |
0.85 |
| chr12_71229100_71229280 | 0.18 |
Gm19045 |
predicted gene, 19045 |
23746 |
0.15 |
| chr15_3581853_3582700 | 0.18 |
Ghr |
growth hormone receptor |
320 |
0.92 |
| chr7_16581555_16581911 | 0.18 |
Gm29443 |
predicted gene 29443 |
32091 |
0.08 |
| chr5_103426807_103426958 | 0.18 |
Ptpn13 |
protein tyrosine phosphatase, non-receptor type 13 |
1381 |
0.47 |
| chr12_113185913_113186788 | 0.18 |
Tmem121 |
transmembrane protein 121 |
420 |
0.74 |
| chr3_87172043_87172207 | 0.18 |
Kirrel |
kirre like nephrin family adhesion molecule 1 |
2425 |
0.26 |
| chr3_82073600_82074661 | 0.18 |
Gucy1b1 |
guanylate cyclase 1, soluble, beta 1 |
543 |
0.77 |
| chr18_35034038_35034310 | 0.18 |
Gm36037 |
predicted gene, 36037 |
51204 |
0.09 |
| chr17_46444584_46445824 | 0.18 |
Gm5093 |
predicted gene 5093 |
5107 |
0.11 |
| chr11_72406421_72408105 | 0.18 |
Smtnl2 |
smoothelin-like 2 |
4155 |
0.15 |
| chr15_61770777_61771058 | 0.18 |
D030024E09Rik |
RIKEN cDNA D030024E09 gene |
3534 |
0.33 |
| chr7_3367594_3368044 | 0.18 |
Gm44257 |
predicted gene, 44257 |
22261 |
0.07 |
| chr10_66901546_66901931 | 0.18 |
Gm47903 |
predicted gene, 47903 |
10126 |
0.15 |
| chr2_113217092_113217461 | 0.18 |
Ryr3 |
ryanodine receptor 3 |
180 |
0.96 |
| chrX_99044012_99044163 | 0.18 |
Stard8 |
START domain containing 8 |
103 |
0.97 |
| chr9_119982824_119983895 | 0.18 |
1700019L13Rik |
RIKEN cDNA 1700019L13 gene |
80 |
0.79 |
| chr17_71782384_71783600 | 0.18 |
Clip4 |
CAP-GLY domain containing linker protein family, member 4 |
375 |
0.84 |
| chr6_47044703_47045103 | 0.18 |
Cntnap2 |
contactin associated protein-like 2 |
157 |
0.64 |
| chr5_134746440_134747928 | 0.18 |
Eln |
elastin |
62 |
0.61 |
| chr9_64014385_64014870 | 0.18 |
Smad6 |
SMAD family member 6 |
2360 |
0.24 |
| chr7_101394207_101396000 | 0.18 |
Arap1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
448 |
0.73 |
| chr4_97776884_97777081 | 0.17 |
Nfia |
nuclear factor I/A |
626 |
0.6 |
| chr3_141491675_141491844 | 0.17 |
Unc5c |
unc-5 netrin receptor C |
26088 |
0.21 |
| chr2_167542817_167543436 | 0.17 |
Snai1 |
snail family zinc finger 1 |
4931 |
0.13 |
| chr9_48986724_48987891 | 0.17 |
Usp28 |
ubiquitin specific peptidase 28 |
1842 |
0.32 |
| chr1_128592817_128593419 | 0.17 |
Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
825 |
0.66 |
| chr6_118479444_118479781 | 0.17 |
Zfp9 |
zinc finger protein 9 |
292 |
0.88 |
| chr5_124529436_124529809 | 0.17 |
Gm43034 |
predicted gene 43034 |
1511 |
0.21 |
| chr8_110805706_110805934 | 0.17 |
Il34 |
interleukin 34 |
52 |
0.67 |
| chr18_37172243_37172684 | 0.17 |
Gm38097 |
predicted gene, 38097 |
101 |
0.95 |
| chr9_71591283_71592381 | 0.17 |
Myzap |
myocardial zonula adherens protein |
433 |
0.84 |
| chr11_105292145_105293042 | 0.17 |
Mrc2 |
mannose receptor, C type 2 |
50 |
0.98 |
| chr10_76583951_76585206 | 0.17 |
Ftcd |
formiminotransferase cyclodeaminase |
2555 |
0.18 |
| chr11_114479225_114479452 | 0.17 |
4932435O22Rik |
RIKEN cDNA 4932435O22 gene |
30478 |
0.2 |
| chr13_48662866_48663051 | 0.17 |
Barx1 |
BarH-like homeobox 1 |
40 |
0.97 |
| chr11_70232254_70232932 | 0.17 |
Mir497 |
microRNA 497 |
2124 |
0.1 |
| chr2_74681727_74681878 | 0.17 |
Hoxd11 |
homeobox D11 |
521 |
0.45 |
| chr14_51738531_51739887 | 0.17 |
Gm49261 |
predicted gene, 49261 |
1896 |
0.19 |
| chr9_89497628_89497779 | 0.17 |
Gm47403 |
predicted gene, 47403 |
62681 |
0.11 |
| chr4_48407474_48407947 | 0.17 |
Invs |
inversin |
25522 |
0.19 |
| chr7_126248023_126249285 | 0.17 |
Sbk1 |
SH3-binding kinase 1 |
208 |
0.69 |
| chr15_42962907_42963107 | 0.17 |
Rspo2 |
R-spondin 2 |
207811 |
0.02 |
| chr13_31810556_31811961 | 0.17 |
Foxc1 |
forkhead box C1 |
4625 |
0.19 |
| chr17_26847878_26848155 | 0.17 |
Nkx2-5 |
NK2 homeobox 5 |
3007 |
0.15 |
| chr6_94281635_94282188 | 0.17 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
1114 |
0.58 |
| chr1_193508615_193509882 | 0.17 |
Mir205hg |
Mir205 host gene |
926 |
0.47 |
| chr6_112273463_112275203 | 0.16 |
Lmcd1 |
LIM and cysteine-rich domains 1 |
575 |
0.71 |
| chr2_94263215_94263820 | 0.16 |
Mir670hg |
MIR670 host gene (non-protein coding) |
1091 |
0.4 |
| chr7_100924536_100925334 | 0.16 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
7172 |
0.15 |
| chr13_3709021_3709715 | 0.16 |
Gm47695 |
predicted gene, 47695 |
50457 |
0.09 |
| chr10_75174015_75174616 | 0.16 |
Bcr |
BCR activator of RhoGEF and GTPase |
350 |
0.89 |
| chr2_71367675_71367887 | 0.16 |
Slc25a12 |
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
32 |
0.97 |
| chr1_12691898_12692734 | 0.16 |
Sulf1 |
sulfatase 1 |
7 |
0.98 |
| chr16_16217315_16217547 | 0.16 |
Pkp2 |
plakophilin 2 |
4113 |
0.23 |
| chr2_35557994_35559313 | 0.16 |
Gm13446 |
predicted gene 13446 |
49 |
0.85 |
| chr2_35621752_35623107 | 0.16 |
Dab2ip |
disabled 2 interacting protein |
269 |
0.93 |
| chr7_104314930_104315471 | 0.16 |
Trim12a |
tripartite motif-containing 12A |
143 |
0.53 |
| chr10_68282341_68283288 | 0.16 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
4074 |
0.25 |
| chr19_47313387_47314780 | 0.16 |
Sh3pxd2a |
SH3 and PX domains 2A |
668 |
0.69 |
| chr7_104352827_104353391 | 0.16 |
Trim12c |
tripartite motif-containing 12C |
229 |
0.84 |
| chr6_114918735_114919212 | 0.16 |
Vgll4 |
vestigial like family member 4 |
2843 |
0.31 |
| chr7_45215829_45217292 | 0.16 |
Tead2 |
TEA domain family member 2 |
113 |
0.88 |
| chr17_26451915_26452093 | 0.16 |
Gm50269 |
predicted gene, 50269 |
12153 |
0.13 |
| chr15_85670939_85672357 | 0.16 |
Lncppara |
long noncoding RNA near Ppara |
18032 |
0.13 |
| chr11_103096614_103097909 | 0.16 |
Plcd3 |
phospholipase C, delta 3 |
4330 |
0.13 |
| chr9_77917044_77917252 | 0.16 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
216 |
0.92 |
| chr7_4520460_4522358 | 0.16 |
Tnni3 |
troponin I, cardiac 3 |
842 |
0.35 |
| chr6_14902200_14902426 | 0.16 |
Foxp2 |
forkhead box P2 |
835 |
0.77 |
| chr10_62338281_62338607 | 0.16 |
Hk1os |
hexokinase 1, opposite strand |
1812 |
0.27 |
| chr2_153224755_153225797 | 0.16 |
Tspyl3 |
TSPY-like 3 |
165 |
0.93 |
| chr5_142819565_142820054 | 0.16 |
Tnrc18 |
trinucleotide repeat containing 18 |
2147 |
0.28 |
| chr5_91402281_91403314 | 0.16 |
Btc |
betacellulin, epidermal growth factor family member |
25 |
0.99 |
| chr11_85833878_85836704 | 0.15 |
Tbx2 |
T-box 2 |
2740 |
0.17 |
| chr4_148001445_148002010 | 0.15 |
Nppa |
natriuretic peptide type A |
1005 |
0.33 |
| chr4_152096248_152097724 | 0.15 |
Plekhg5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
267 |
0.86 |
| chr1_135788611_135788952 | 0.15 |
Tnni1 |
troponin I, skeletal, slow 1 |
5716 |
0.15 |
| chr5_103693169_103693605 | 0.15 |
Aff1 |
AF4/FMR2 family, member 1 |
1013 |
0.5 |
| chr7_75141792_75142655 | 0.15 |
Sv2b |
synaptic vesicle glycoprotein 2 b |
5503 |
0.3 |
| chr7_19747051_19747276 | 0.15 |
Nectin2 |
nectin cell adhesion molecule 2 |
2370 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.2 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.4 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.2 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 0.3 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.3 | GO:2000618 | regulation of histone H4-K16 acetylation(GO:2000618) |
| 0.1 | 0.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.2 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.1 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.2 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.0 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.0 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.0 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.0 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.3 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0018638 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.4 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |