Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
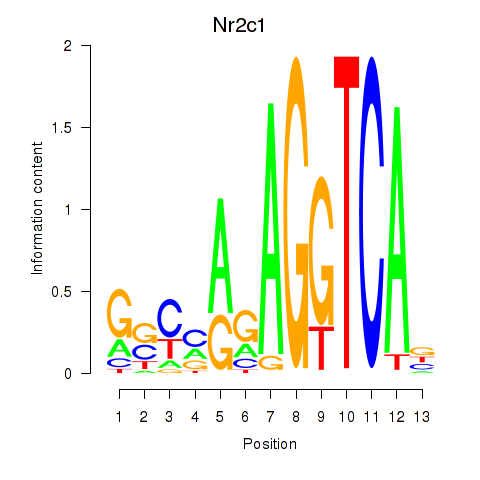
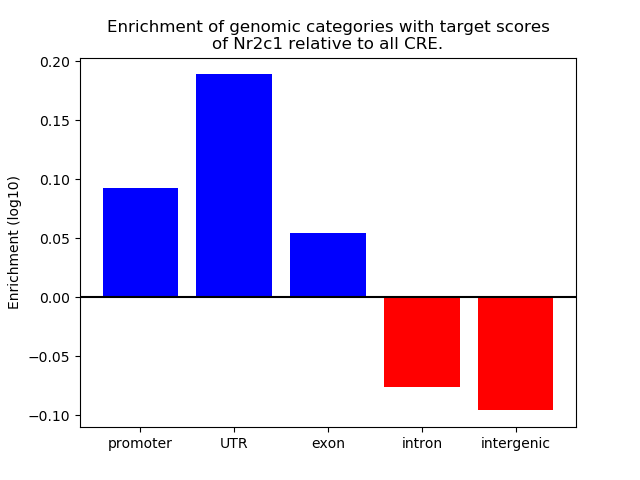
Results for Nr2c1
Z-value: 0.59
Transcription factors associated with Nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c1
|
ENSMUSG00000005897.8 | nuclear receptor subfamily 2, group C, member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_94149504_94150012 | Nr2c1 | 1339 | 0.351064 | -0.28 | 2.9e-02 | Click! |
| chr10_94148808_94148959 | Nr2c1 | 464 | 0.755356 | -0.08 | 5.4e-01 | Click! |
| chr10_94150806_94150977 | Nr2c1 | 2472 | 0.212914 | -0.08 | 5.6e-01 | Click! |
| chr10_94147624_94148712 | Nr2c1 | 145 | 0.941655 | 0.04 | 7.5e-01 | Click! |
| chr10_94165488_94165639 | Nr2c1 | 9277 | 0.140757 | 0.04 | 7.7e-01 | Click! |
Activity of the Nr2c1 motif across conditions
Conditions sorted by the z-value of the Nr2c1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_144547334_144548656 | 1.40 |
Nptx2 |
neuronal pentraxin 2 |
2093 |
0.41 |
| chr9_120021605_120021794 | 1.18 |
Xirp1 |
xin actin-binding repeat containing 1 |
1884 |
0.19 |
| chr18_76855816_76857376 | 1.16 |
Skor2 |
SKI family transcriptional corepressor 2 |
191 |
0.96 |
| chr7_78575006_78575482 | 1.14 |
Gm39038 |
predicted gene, 39038 |
12 |
0.97 |
| chrX_170674573_170675954 | 1.11 |
Asmt |
acetylserotonin O-methyltransferase |
2619 |
0.41 |
| chr9_87739615_87740838 | 1.10 |
Gm37374 |
predicted gene, 37374 |
206 |
0.93 |
| chr2_83758891_83759726 | 0.99 |
Gm13684 |
predicted gene 13684 |
557 |
0.73 |
| chr5_34182980_34183403 | 0.92 |
Mxd4 |
Max dimerization protein 4 |
1179 |
0.3 |
| chr11_55604950_55605292 | 0.91 |
Glra1 |
glycine receptor, alpha 1 subunit |
2612 |
0.3 |
| chr10_41070697_41071486 | 0.90 |
Gpr6 |
G protein-coupled receptor 6 |
1194 |
0.41 |
| chr13_53469792_53470715 | 0.88 |
Msx2 |
msh homeobox 2 |
2821 |
0.27 |
| chr17_43954528_43954986 | 0.86 |
Rcan2 |
regulator of calcineurin 2 |
1506 |
0.53 |
| chr5_113490881_113491656 | 0.85 |
Wscd2 |
WSC domain containing 2 |
516 |
0.83 |
| chr6_82091782_82092469 | 0.82 |
Gm15864 |
predicted gene 15864 |
39544 |
0.14 |
| chr4_153481347_153482221 | 0.82 |
Ajap1 |
adherens junction associated protein 1 |
395 |
0.92 |
| chr4_141010412_141011785 | 0.81 |
Mfap2 |
microfibrillar-associated protein 2 |
454 |
0.71 |
| chr14_122459815_122460898 | 0.80 |
Zic5 |
zinc finger protein of the cerebellum 5 |
335 |
0.81 |
| chr1_194622071_194623282 | 0.80 |
Plxna2 |
plexin A2 |
2851 |
0.26 |
| chr9_121299469_121300553 | 0.79 |
Trak1 |
trafficking protein, kinesin binding 1 |
2182 |
0.23 |
| chr5_24334809_24334960 | 0.78 |
Gm15589 |
predicted gene 15589 |
961 |
0.39 |
| chr9_41376046_41377501 | 0.76 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
212 |
0.93 |
| chr1_95662771_95663208 | 0.76 |
St8sia4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
4546 |
0.23 |
| chr8_108406602_108407498 | 0.76 |
Gm39244 |
predicted gene, 39244 |
129897 |
0.05 |
| chr18_31443497_31444446 | 0.73 |
Syt4 |
synaptotagmin IV |
3435 |
0.19 |
| chr7_99272446_99273539 | 0.72 |
Map6 |
microtubule-associated protein 6 |
3860 |
0.15 |
| chr10_79806462_79808470 | 0.71 |
Palm |
paralemmin |
785 |
0.35 |
| chr10_81021201_81021561 | 0.69 |
Gm16099 |
predicted gene 16099 |
3430 |
0.11 |
| chr7_46097493_46097815 | 0.68 |
Kcnj11 |
potassium inwardly rectifying channel, subfamily J, member 11 |
772 |
0.46 |
| chr14_25586887_25588082 | 0.67 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
19873 |
0.17 |
| chr15_89532557_89533956 | 0.67 |
Shank3 |
SH3 and multiple ankyrin repeat domains 3 |
366 |
0.78 |
| chr15_73839069_73840200 | 0.66 |
Mroh5 |
maestro heat-like repeat family member 5 |
37 |
0.97 |
| chr15_103026302_103028215 | 0.65 |
Hoxc4 |
homeobox C4 |
7137 |
0.09 |
| chr5_142811476_142811629 | 0.64 |
Tnrc18 |
trinucleotide repeat containing 18 |
5828 |
0.19 |
| chr2_158375202_158376961 | 0.64 |
Snhg11 |
small nucleolar RNA host gene 11 |
319 |
0.74 |
| chr17_35505957_35506108 | 0.64 |
Pou5f1 |
POU domain, class 5, transcription factor 1 |
14 |
0.93 |
| chr8_125897868_125898882 | 0.63 |
Pcnx2 |
pecanex homolog 2 |
58 |
0.88 |
| chr8_47284401_47285223 | 0.63 |
Stox2 |
storkhead box 2 |
4550 |
0.27 |
| chr16_6841576_6842071 | 0.63 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
32601 |
0.26 |
| chr7_49914124_49915124 | 0.61 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
2924 |
0.34 |
| chr9_124441736_124442440 | 0.61 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
658 |
0.65 |
| chr7_128205575_128206445 | 0.61 |
Cox6a2 |
cytochrome c oxidase subunit 6A2 |
377 |
0.67 |
| chr18_36939256_36940257 | 0.60 |
Pcdha2 |
protocadherin alpha 2 |
532 |
0.46 |
| chr3_152980951_152982389 | 0.60 |
St6galnac5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
382 |
0.86 |
| chr11_59138017_59138780 | 0.59 |
Obscn |
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
772 |
0.52 |
| chr4_151127830_151127981 | 0.59 |
Camta1 |
calmodulin binding transcription activator 1 |
1590 |
0.42 |
| chr2_158609356_158610186 | 0.59 |
Gm14204 |
predicted gene 14204 |
819 |
0.36 |
| chr8_12947702_12949640 | 0.59 |
Mcf2l |
mcf.2 transforming sequence-like |
718 |
0.37 |
| chr7_105785466_105785617 | 0.58 |
Dchs1 |
dachsous cadherin related 1 |
2011 |
0.18 |
| chr8_62952744_62952895 | 0.58 |
Spock3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
894 |
0.63 |
| chr7_27957807_27957958 | 0.58 |
Gm10651 |
predicted pseudogene 10651 |
8119 |
0.11 |
| chr8_22508392_22509319 | 0.58 |
Slc20a2 |
solute carrier family 20, member 2 |
2629 |
0.22 |
| chr9_124439879_124441093 | 0.58 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
382 |
0.83 |
| chr13_64035278_64035429 | 0.57 |
Hsd17b3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
27780 |
0.16 |
| chr9_110051810_110053856 | 0.57 |
Map4 |
microtubule-associated protein 4 |
781 |
0.54 |
| chr3_88210226_88210408 | 0.56 |
Gm3764 |
predicted gene 3764 |
845 |
0.35 |
| chr9_91352783_91353195 | 0.56 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
2036 |
0.19 |
| chrX_110811626_110812334 | 0.56 |
Gm44593 |
predicted gene 44593 |
344 |
0.89 |
| chr15_31296980_31298102 | 0.55 |
Ankrd33b |
ankyrin repeat domain 33B |
10653 |
0.14 |
| chr11_9118055_9119445 | 0.54 |
Upp1 |
uridine phosphorylase 1 |
37 |
0.98 |
| chr13_94876260_94878233 | 0.54 |
Otp |
orthopedia homeobox |
1644 |
0.4 |
| chr8_123894797_123895120 | 0.54 |
Acta1 |
actin, alpha 1, skeletal muscle |
207 |
0.85 |
| chr11_3504903_3505132 | 0.54 |
Inpp5j |
inositol polyphosphate 5-phosphatase J |
196 |
0.87 |
| chr2_26339437_26339737 | 0.53 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
139 |
0.91 |
| chr9_117837070_117837361 | 0.53 |
Rbms3 |
RNA binding motif, single stranded interacting protein |
35369 |
0.18 |
| chr12_106080982_106081684 | 0.53 |
Gm46378 |
predicted gene, 46378 |
3037 |
0.25 |
| chr14_24616899_24617392 | 0.52 |
4930542C16Rik |
RIKEN cDNA 4930542C16 gene |
159 |
0.78 |
| chr16_31943940_31944452 | 0.52 |
Pigz |
phosphatidylinositol glycan anchor biosynthesis, class Z |
2283 |
0.13 |
| chr17_55970296_55971683 | 0.52 |
Shd |
src homology 2 domain-containing transforming protein D |
400 |
0.68 |
| chr10_80009830_80009981 | 0.51 |
Arhgap45 |
Rho GTPase activating protein 45 |
6748 |
0.07 |
| chr2_32605823_32605974 | 0.51 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
1063 |
0.28 |
| chr13_83715222_83716973 | 0.51 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
5284 |
0.15 |
| chr5_115431565_115432258 | 0.51 |
Msi1 |
musashi RNA-binding protein 1 |
1306 |
0.22 |
| chr3_17798321_17798878 | 0.51 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
2855 |
0.23 |
| chr1_134339239_134339390 | 0.50 |
Ppfia4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
6386 |
0.13 |
| chr6_52965291_52965504 | 0.50 |
Gm26215 |
predicted gene, 26215 |
45135 |
0.14 |
| chr7_79504311_79505700 | 0.50 |
Mir9-3 |
microRNA 9-3 |
259 |
0.82 |
| chr2_170734259_170734932 | 0.50 |
Dok5 |
docking protein 5 |
2788 |
0.35 |
| chr12_118852808_118853969 | 0.50 |
Sp8 |
trans-acting transcription factor 8 |
5802 |
0.24 |
| chr6_83404978_83405260 | 0.49 |
Mir6374 |
microRNA 6374 |
4017 |
0.14 |
| chr12_76081146_76082447 | 0.49 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
140 |
0.97 |
| chr10_76344505_76344665 | 0.49 |
Dip2a |
disco interacting protein 2 homolog A |
647 |
0.64 |
| chr3_105706804_105707390 | 0.48 |
Inka2 |
inka box actin regulator 2 |
1639 |
0.26 |
| chr2_26634084_26634235 | 0.48 |
Dipk1b |
divergent protein kinase domain 1B |
44 |
0.94 |
| chr5_23429149_23429300 | 0.48 |
5031425E22Rik |
RIKEN cDNA 5031425E22 gene |
4085 |
0.16 |
| chr5_102100207_102100695 | 0.48 |
Gm29707 |
predicted gene, 29707 |
29601 |
0.14 |
| chr1_172018523_172018674 | 0.47 |
Vangl2 |
VANGL planar cell polarity 2 |
8113 |
0.14 |
| chr17_45920036_45920187 | 0.47 |
Gm41584 |
predicted gene, 41584 |
34765 |
0.12 |
| chr12_105453821_105454975 | 0.47 |
D430019H16Rik |
RIKEN cDNA D430019H16 gene |
542 |
0.76 |
| chr7_79498955_79500626 | 0.47 |
Mir9-3hg |
Mir9-3 host gene |
236 |
0.84 |
| chr2_136713069_136714459 | 0.47 |
Snap25 |
synaptosomal-associated protein 25 |
286 |
0.92 |
| chr2_113327774_113328199 | 0.46 |
Fmn1 |
formin 1 |
204 |
0.94 |
| chr5_134101487_134102203 | 0.46 |
Castor2 |
cytosolic arginine sensor for mTORC1 subunit 2 |
1840 |
0.26 |
| chr4_154945869_154946537 | 0.46 |
Hes5 |
hes family bHLH transcription factor 5 |
14720 |
0.11 |
| chr17_8429499_8430158 | 0.46 |
T |
brachyury, T-box transcription factor T |
4595 |
0.19 |
| chr5_112520948_112521383 | 0.45 |
Sez6l |
seizure related 6 homolog like |
46038 |
0.1 |
| chr13_34133417_34134151 | 0.45 |
Gm36500 |
predicted gene, 36500 |
76 |
0.95 |
| chr1_132316080_132316995 | 0.45 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
372 |
0.79 |
| chr16_77417791_77418279 | 0.45 |
Gm38071 |
predicted gene, 38071 |
1411 |
0.28 |
| chr7_25068107_25069005 | 0.45 |
Grik5 |
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
3363 |
0.14 |
| chr4_22478508_22478806 | 0.45 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
9709 |
0.17 |
| chr2_163206219_163206674 | 0.45 |
Tox2 |
TOX high mobility group box family member 2 |
1893 |
0.35 |
| chr12_12693706_12694681 | 0.44 |
Gm25538 |
predicted gene, 25538 |
5846 |
0.17 |
| chr8_23132120_23132667 | 0.44 |
Gm15815 |
predicted gene 15815 |
4937 |
0.11 |
| chr2_167537717_167538031 | 0.44 |
Snai1 |
snail family zinc finger 1 |
321 |
0.81 |
| chr7_139086002_139087206 | 0.44 |
Dpysl4 |
dihydropyrimidinase-like 4 |
576 |
0.74 |
| chr2_30097085_30097294 | 0.44 |
Zdhhc12 |
zinc finger, DHHC domain containing 12 |
3541 |
0.12 |
| chr2_6883618_6884699 | 0.44 |
Gm13389 |
predicted gene 13389 |
112 |
0.85 |
| chrX_99137870_99138145 | 0.44 |
Efnb1 |
ephrin B1 |
124 |
0.97 |
| chr13_99513069_99513554 | 0.43 |
Map1b |
microtubule-associated protein 1B |
3207 |
0.2 |
| chr18_79071481_79071862 | 0.43 |
Setbp1 |
SET binding protein 1 |
37720 |
0.22 |
| chr1_159524626_159524870 | 0.43 |
Tnr |
tenascin R |
907 |
0.65 |
| chr3_89763591_89764832 | 0.43 |
Chrnb2 |
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
397 |
0.76 |
| chr8_87037668_87038705 | 0.43 |
Rps13-ps1 |
ribosomal protein S13, pseudogene 1 |
9503 |
0.17 |
| chr5_136238361_136238512 | 0.43 |
Sh2b2 |
SH2B adaptor protein 2 |
6401 |
0.14 |
| chr16_60602088_60602266 | 0.43 |
Gm9017 |
predicted gene 9017 |
2589 |
0.24 |
| chr11_5802324_5803834 | 0.43 |
Pgam2 |
phosphoglycerate mutase 2 |
654 |
0.56 |
| chr7_44973695_44975193 | 0.43 |
Cpt1c |
carnitine palmitoyltransferase 1c |
331 |
0.61 |
| chr3_95696412_95696933 | 0.43 |
Adamtsl4 |
ADAMTS-like 4 |
8755 |
0.1 |
| chr13_64315557_64315911 | 0.42 |
Prxl2c |
peroxiredoxin like 2C |
3024 |
0.13 |
| chr5_24982764_24982915 | 0.42 |
1500035N22Rik |
RIKEN cDNA 1500035N22 gene |
3003 |
0.24 |
| chr7_78575992_78576673 | 0.42 |
Gm39038 |
predicted gene, 39038 |
1100 |
0.39 |
| chr5_134238122_134238273 | 0.42 |
Gtf2i |
general transcription factor II I |
2848 |
0.18 |
| chr18_37663804_37664034 | 0.41 |
Gm37118 |
predicted gene, 37118 |
1287 |
0.14 |
| chr8_120575933_120576084 | 0.41 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
4679 |
0.11 |
| chr18_59062200_59063436 | 0.41 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
307 |
0.94 |
| chr18_67511489_67511640 | 0.41 |
Spire1 |
spire type actin nucleation factor 1 |
5830 |
0.18 |
| chr5_111424407_111425623 | 0.41 |
Gm43119 |
predicted gene 43119 |
1426 |
0.38 |
| chr5_110545528_110545748 | 0.41 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
1283 |
0.39 |
| chr11_120020575_120020893 | 0.40 |
Aatk |
apoptosis-associated tyrosine kinase |
566 |
0.59 |
| chr18_84074730_84075170 | 0.40 |
Tshz1 |
teashirt zinc finger family member 1 |
10125 |
0.16 |
| chr8_95354616_95354905 | 0.40 |
Mmp15 |
matrix metallopeptidase 15 |
2492 |
0.19 |
| chr13_81944263_81944829 | 0.40 |
F830210D05Rik |
RIKEN cDNA F830210D05 gene |
14859 |
0.26 |
| chr1_194621653_194621902 | 0.40 |
Plxna2 |
plexin A2 |
1952 |
0.34 |
| chr12_8495918_8496727 | 0.40 |
Rhob |
ras homolog family member B |
3687 |
0.18 |
| chr4_135174988_135175978 | 0.39 |
Runx3 |
runt related transcription factor 3 |
21047 |
0.15 |
| chr3_156564902_156565177 | 0.39 |
Negr1 |
neuronal growth regulator 1 |
2889 |
0.27 |
| chr6_48537635_48538183 | 0.39 |
Atp6v0e2 |
ATPase, H+ transporting, lysosomal V0 subunit E2 |
244 |
0.61 |
| chr8_71685721_71686960 | 0.38 |
Insl3 |
insulin-like 3 |
2874 |
0.12 |
| chr10_13982681_13983672 | 0.38 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
16152 |
0.19 |
| chr3_84217272_84217435 | 0.38 |
Trim2 |
tripartite motif-containing 2 |
3492 |
0.28 |
| chr5_118929922_118930545 | 0.38 |
Gm43342 |
predicted gene 43342 |
2110 |
0.32 |
| chr10_80177626_80177980 | 0.38 |
Efna2 |
ephrin A2 |
1679 |
0.18 |
| chr12_111353800_111353990 | 0.38 |
Cdc42bpb |
CDC42 binding protein kinase beta |
23724 |
0.13 |
| chr4_33716448_33717151 | 0.38 |
Gm24341 |
predicted gene, 24341 |
35308 |
0.2 |
| chr9_57696029_57697476 | 0.38 |
Cyp1a1 |
cytochrome P450, family 1, subfamily a, polypeptide 1 |
861 |
0.48 |
| chr2_168447547_168447953 | 0.38 |
Gm14234 |
predicted gene 14234 |
29176 |
0.19 |
| chr11_55601469_55602112 | 0.38 |
Glra1 |
glycine receptor, alpha 1 subunit |
5943 |
0.22 |
| chr19_4758474_4759201 | 0.38 |
Rbm4b |
RNA binding motif protein 4B |
1932 |
0.18 |
| chr11_102394791_102396113 | 0.38 |
Rundc3a |
RUN domain containing 3A |
2049 |
0.17 |
| chr13_72298974_72299125 | 0.37 |
Gm4052 |
predicted gene 4052 |
51172 |
0.15 |
| chr1_89457566_89457902 | 0.37 |
Agap1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
2478 |
0.29 |
| chr1_135374257_135375340 | 0.37 |
Shisa4 |
shisa family member 4 |
279 |
0.86 |
| chr3_96424839_96424993 | 0.37 |
Gm24136 |
predicted gene, 24136 |
2183 |
0.1 |
| chr15_103038687_103039944 | 0.37 |
Hoxc4 |
homeobox C4 |
4920 |
0.1 |
| chr4_126464266_126464731 | 0.37 |
Ago1 |
argonaute RISC catalytic subunit 1 |
2716 |
0.17 |
| chr7_45304558_45305606 | 0.37 |
Trpm4 |
transient receptor potential cation channel, subfamily M, member 4 |
122 |
0.9 |
| chr12_106474400_106475045 | 0.37 |
Gm3191 |
predicted gene 3191 |
15572 |
0.17 |
| chr3_3507690_3508493 | 0.37 |
Hnf4g |
hepatocyte nuclear factor 4, gamma |
61 |
0.98 |
| chr17_12762808_12762986 | 0.37 |
Igf2r |
insulin-like growth factor 2 receptor |
6767 |
0.13 |
| chr3_105962812_105962963 | 0.37 |
Wdr77 |
WD repeat domain 77 |
1265 |
0.24 |
| chr1_69790500_69791180 | 0.36 |
Gm37930 |
predicted gene, 37930 |
19972 |
0.21 |
| chr5_132394811_132395242 | 0.36 |
Gm42989 |
predicted gene 42989 |
22345 |
0.15 |
| chr4_149075794_149076058 | 0.35 |
Gm9506 |
predicted gene 9506 |
20457 |
0.12 |
| chr1_172218328_172218648 | 0.35 |
Casq1 |
calsequestrin 1 |
1227 |
0.28 |
| chr2_119268931_119270073 | 0.35 |
Rhov |
ras homolog family member V |
1770 |
0.21 |
| chr19_53793336_53793637 | 0.35 |
Rbm20 |
RNA binding motif protein 20 |
178 |
0.94 |
| chr10_8761475_8762950 | 0.35 |
Sash1 |
SAM and SH3 domain containing 1 |
262 |
0.93 |
| chr9_32835753_32835904 | 0.35 |
Gm37167 |
predicted gene, 37167 |
4094 |
0.24 |
| chr12_55438453_55439133 | 0.35 |
Psma6 |
proteasome subunit alpha 6 |
31410 |
0.13 |
| chr2_49619321_49620607 | 0.35 |
Kif5c |
kinesin family member 5C |
666 |
0.78 |
| chr6_136167149_136168437 | 0.34 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
4096 |
0.25 |
| chr17_35140108_35141075 | 0.34 |
Bag6 |
BCL2-associated athanogene 6 |
360 |
0.57 |
| chr11_98360332_98361761 | 0.34 |
Stard3 |
START domain containing 3 |
2602 |
0.12 |
| chr11_3910058_3910209 | 0.34 |
Slc35e4 |
solute carrier family 35, member E4 |
4531 |
0.11 |
| chr1_66467636_66468234 | 0.34 |
Unc80 |
unc-80, NALCN activator |
432 |
0.85 |
| chr9_72665710_72665959 | 0.34 |
Nedd4 |
neural precursor cell expressed, developmentally down-regulated 4 |
2835 |
0.14 |
| chr6_5725575_5726689 | 0.34 |
Dync1i1 |
dynein cytoplasmic 1 intermediate chain 1 |
314 |
0.94 |
| chr11_117436519_117437340 | 0.34 |
Gm11732 |
predicted gene 11732 |
10751 |
0.14 |
| chr10_128381063_128381470 | 0.34 |
Mir6914 |
microRNA 6914 |
1572 |
0.14 |
| chrX_105390628_105392456 | 0.33 |
5330434G04Rik |
RIKEN cDNA 5330434G04 gene |
212 |
0.93 |
| chr3_109286535_109286686 | 0.33 |
Vav3 |
vav 3 oncogene |
54043 |
0.14 |
| chr3_34656299_34657721 | 0.33 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
974 |
0.4 |
| chr2_29046202_29046836 | 0.33 |
Cfap77 |
cilia and flagella associated protein 77 |
7159 |
0.17 |
| chr9_108824114_108825614 | 0.33 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr17_37002443_37003493 | 0.33 |
Zfp57 |
zinc finger protein 57 |
70 |
0.93 |
| chr5_117068973_117069842 | 0.33 |
Gm7478 |
predicted gene 7478 |
10595 |
0.17 |
| chr2_163392737_163392895 | 0.33 |
Jph2 |
junctophilin 2 |
5133 |
0.15 |
| chr7_65157281_65157432 | 0.33 |
Fam189a1 |
family with sequence similarity 189, member A1 |
786 |
0.63 |
| chr17_80148180_80148718 | 0.33 |
Galm |
galactose mutarotase |
3297 |
0.21 |
| chr8_125349587_125349941 | 0.33 |
Gm16237 |
predicted gene 16237 |
122441 |
0.05 |
| chr13_48155311_48155462 | 0.32 |
Gm36346 |
predicted gene, 36346 |
81865 |
0.08 |
| chr11_101471331_101471482 | 0.32 |
Rnd2 |
Rho family GTPase 2 |
3231 |
0.09 |
| chrX_41404646_41404797 | 0.32 |
Gria3 |
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
1981 |
0.42 |
| chr4_82510609_82512039 | 0.32 |
Gm11266 |
predicted gene 11266 |
3308 |
0.25 |
| chr19_4440980_4441188 | 0.32 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
1525 |
0.24 |
| chr8_87838025_87838279 | 0.32 |
Zfp423 |
zinc finger protein 423 |
33713 |
0.22 |
| chr19_16873202_16873655 | 0.32 |
Foxb2 |
forkhead box B2 |
377 |
0.85 |
| chr7_34569925_34571186 | 0.32 |
Gm12784 |
predicted gene 12784 |
23349 |
0.15 |
| chr12_71048832_71049275 | 0.32 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
712 |
0.65 |
| chr8_71377836_71378740 | 0.32 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
2498 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 0.5 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.2 | 0.8 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.4 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.4 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.1 | 0.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 1.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.2 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.5 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.4 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.1 | 0.3 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.2 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.4 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 0.4 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.2 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0052151 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0003284 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.0 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0098828 | modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0009180 | purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.0 | GO:1902659 | pharyngeal arch artery morphogenesis(GO:0061626) regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.0 | GO:1903365 | regulation of fear response(GO:1903365) positive regulation of fear response(GO:1903367) regulation of behavioral fear response(GO:2000822) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.3 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.9 | GO:0008306 | associative learning(GO:0008306) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 | 0.2 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0060770 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.0 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.0 | GO:1900238 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.2 | GO:0043153 | photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.0 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.0 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0001771 | immunological synapse formation(GO:0001771) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.8 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0015922 | aspartate oxidase activity(GO:0015922) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.4 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |