Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
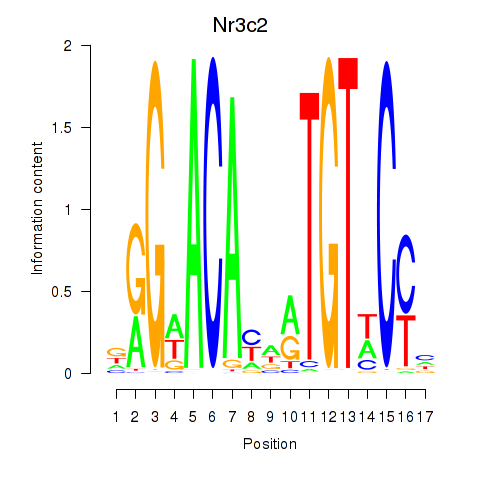
Results for Nr3c2
Z-value: 0.75
Transcription factors associated with Nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr3c2
|
ENSMUSG00000031618.7 | nuclear receptor subfamily 3, group C, member 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_77132765_77133214 | Nr3c2 | 4976 | 0.232146 | 0.51 | 2.9e-05 | Click! |
| chr8_77044923_77045251 | Nr3c2 | 82926 | 0.088938 | 0.41 | 1.3e-03 | Click! |
| chr8_76902707_76902893 | Nr3c2 | 324 | 0.882397 | 0.39 | 2.0e-03 | Click! |
| chr8_76912612_76912763 | Nr3c2 | 4415 | 0.245749 | 0.33 | 9.5e-03 | Click! |
| chr8_76901608_76902608 | Nr3c2 | 180 | 0.933412 | -0.23 | 8.2e-02 | Click! |
Activity of the Nr3c2 motif across conditions
Conditions sorted by the z-value of the Nr3c2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_101448217_101448875 | 4.30 |
Ifi35 |
interferon-induced protein 35 |
24 |
0.92 |
| chr5_143625482_143626083 | 3.48 |
Cyth3 |
cytohesin 3 |
3248 |
0.24 |
| chr16_38374654_38375717 | 2.81 |
Popdc2 |
popeye domain containing 2 |
3187 |
0.17 |
| chrX_13469647_13469909 | 2.70 |
Nyx |
nyctalopin |
3668 |
0.23 |
| chr6_34862829_34863673 | 2.66 |
Tmem140 |
transmembrane protein 140 |
11 |
0.96 |
| chr2_158141899_158142209 | 2.46 |
Tgm2 |
transglutaminase 2, C polypeptide |
2318 |
0.24 |
| chr18_12257613_12257833 | 2.16 |
Ankrd29 |
ankyrin repeat domain 29 |
13974 |
0.14 |
| chr12_12488453_12488685 | 2.11 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
95954 |
0.08 |
| chr11_115899671_115901427 | 2.10 |
Smim5 |
small integral membrane protein 5 |
347 |
0.75 |
| chr10_43518568_43518757 | 2.10 |
Gm47889 |
predicted gene, 47889 |
91 |
0.95 |
| chr10_117281690_117282226 | 2.08 |
Lyz2 |
lysozyme 2 |
363 |
0.81 |
| chr8_94178564_94180490 | 2.06 |
Mt1 |
metallothionein 1 |
286 |
0.81 |
| chr4_115828937_115829088 | 2.05 |
Mob3c |
MOB kinase activator 3C |
354 |
0.81 |
| chr12_103630582_103631271 | 1.96 |
Serpina10 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
495 |
0.69 |
| chr6_90624636_90625023 | 1.95 |
Slc41a3 |
solute carrier family 41, member 3 |
5682 |
0.15 |
| chr13_112046443_112047142 | 1.92 |
Gm15323 |
predicted gene 15323 |
41290 |
0.14 |
| chr15_5243498_5243891 | 1.86 |
Ptger4 |
prostaglandin E receptor 4 (subtype EP4) |
478 |
0.75 |
| chr15_36715797_36716780 | 1.81 |
Gm49263 |
predicted gene, 49263 |
7359 |
0.15 |
| chr4_155468952_155469786 | 1.74 |
Tmem52 |
transmembrane protein 52 |
222 |
0.87 |
| chr11_102437215_102438375 | 1.74 |
Fam171a2 |
family with sequence similarity 171, member A2 |
1982 |
0.17 |
| chr16_36934847_36935880 | 1.73 |
Hcls1 |
hematopoietic cell specific Lyn substrate 1 |
380 |
0.77 |
| chr7_97748212_97748509 | 1.70 |
Aqp11 |
aquaporin 11 |
10071 |
0.16 |
| chr11_90686079_90686230 | 1.70 |
Tom1l1 |
target of myb1-like 1 (chicken) |
1425 |
0.46 |
| chr1_165717936_165719081 | 1.68 |
Gm37073 |
predicted gene, 37073 |
3048 |
0.14 |
| chr12_76709422_76710754 | 1.66 |
Sptb |
spectrin beta, erythrocytic |
65 |
0.98 |
| chr1_87650650_87651222 | 1.64 |
Inpp5d |
inositol polyphosphate-5-phosphatase D |
25303 |
0.15 |
| chr2_167671877_167672567 | 1.62 |
Gm14320 |
predicted gene 14320 |
10113 |
0.1 |
| chr15_102102083_102102892 | 1.61 |
Tns2 |
tensin 2 |
259 |
0.86 |
| chr16_21826993_21827376 | 1.60 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
1242 |
0.33 |
| chr4_154024404_154026596 | 1.55 |
Smim1 |
small integral membrane protein 1 |
116 |
0.93 |
| chr19_53194082_53195694 | 1.50 |
Add3 |
adducin 3 (gamma) |
65 |
0.97 |
| chr17_27056910_27058117 | 1.47 |
Itpr3 |
inositol 1,4,5-triphosphate receptor 3 |
209 |
0.87 |
| chr11_69096707_69097996 | 1.47 |
Per1 |
period circadian clock 1 |
1597 |
0.16 |
| chr14_46536264_46536434 | 1.45 |
Gm48912 |
predicted gene, 48912 |
19907 |
0.11 |
| chr2_181079821_181080027 | 1.44 |
Kcnq2 |
potassium voltage-gated channel, subfamily Q, member 2 |
15497 |
0.13 |
| chr8_71406049_71406500 | 1.41 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
250 |
0.83 |
| chr11_34845636_34846358 | 1.40 |
Gm22022 |
predicted gene, 22022 |
3643 |
0.22 |
| chr18_54436599_54436760 | 1.40 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
14384 |
0.22 |
| chr1_191956577_191957080 | 1.36 |
Rd3 |
retinal degeneration 3 |
20542 |
0.11 |
| chr16_34922552_34923302 | 1.30 |
Mylk |
myosin, light polypeptide kinase |
7415 |
0.19 |
| chr3_51661793_51662907 | 1.29 |
Mgst2 |
microsomal glutathione S-transferase 2 |
1125 |
0.38 |
| chr9_22136067_22136415 | 1.27 |
Acp5 |
acid phosphatase 5, tartrate resistant |
530 |
0.54 |
| chr8_111295256_111295437 | 1.27 |
Rfwd3 |
ring finger and WD repeat domain 3 |
4845 |
0.16 |
| chr2_174159770_174159921 | 1.27 |
Gm10714 |
predicted gene 10714 |
24073 |
0.16 |
| chr10_70127596_70128163 | 1.26 |
Ccdc6 |
coiled-coil domain containing 6 |
30758 |
0.2 |
| chr17_27018044_27018864 | 1.26 |
Ggnbp1 |
gametogenetin binding protein 1 |
392 |
0.71 |
| chr10_128743161_128743519 | 1.24 |
Dgka |
diacylglycerol kinase, alpha |
735 |
0.43 |
| chr12_40016524_40017443 | 1.24 |
Arl4a |
ADP-ribosylation factor-like 4A |
2499 |
0.29 |
| chr3_145591307_145591663 | 1.23 |
Znhit6 |
zinc finger, HIT type 6 |
4383 |
0.23 |
| chr11_69915606_69917001 | 1.22 |
Gps2 |
G protein pathway suppressor 2 |
262 |
0.72 |
| chr5_112288747_112288992 | 1.21 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
107 |
0.94 |
| chr4_104913144_104913947 | 1.21 |
Fyb2 |
FYN binding protein 2 |
21 |
0.98 |
| chr9_32342355_32342506 | 1.18 |
Kcnj5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
1807 |
0.31 |
| chr3_52029454_52030701 | 1.17 |
Gm37465 |
predicted gene, 37465 |
26052 |
0.1 |
| chr7_80995833_80996208 | 1.16 |
Gm44967 |
predicted gene 44967 |
1337 |
0.26 |
| chr7_78914724_78915335 | 1.12 |
Isg20 |
interferon-stimulated protein |
700 |
0.57 |
| chr15_6081136_6081368 | 1.10 |
Gm8047 |
predicted gene 8047 |
46291 |
0.16 |
| chr19_5306588_5308182 | 1.09 |
Gm42064 |
predicted gene, 42064 |
31 |
0.93 |
| chr3_89570102_89570404 | 1.09 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
50089 |
0.1 |
| chr1_75144472_75144783 | 1.08 |
Retreg2 |
reticulophagy regulator family member 2 |
677 |
0.43 |
| chr17_47507886_47508650 | 1.08 |
Ccnd3 |
cyclin D3 |
3027 |
0.19 |
| chr16_15889582_15890122 | 1.08 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
1436 |
0.42 |
| chr1_172510177_172510457 | 1.07 |
Gm37125 |
predicted gene, 37125 |
2932 |
0.14 |
| chr8_121904943_121905414 | 1.07 |
Slc7a5 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
2516 |
0.15 |
| chr13_51846753_51848355 | 1.06 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
810 |
0.69 |
| chr8_105831399_105831715 | 1.05 |
Tsnaxip1 |
translin-associated factor X (Tsnax) interacting protein 1 |
3805 |
0.1 |
| chr5_66079108_66079628 | 1.04 |
Rbm47 |
RNA binding motif protein 47 |
1616 |
0.27 |
| chr4_40849281_40849945 | 1.04 |
Mir5123 |
microRNA 5123 |
525 |
0.38 |
| chr1_181255916_181257436 | 1.04 |
Rpl35a-ps2 |
ribosomal protein L35A, pseudogene 2 |
14356 |
0.14 |
| chr19_5559639_5561263 | 1.03 |
1810058N15Rik |
RIKEN cDNA 1810058N15 gene |
191 |
0.47 |
| chr11_44510011_44510178 | 1.01 |
Rnf145 |
ring finger protein 145 |
8870 |
0.17 |
| chr2_30716357_30717429 | 0.99 |
Gm14488 |
predicted gene 14488 |
150 |
0.94 |
| chr14_48133184_48133904 | 0.99 |
Gm49310 |
predicted gene, 49310 |
680 |
0.61 |
| chr13_108670846_108671019 | 0.98 |
Rps3a3 |
ribosomal protein S3A3 |
329 |
0.92 |
| chr17_45599296_45600146 | 0.97 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
115 |
0.92 |
| chr9_21352648_21353105 | 0.96 |
Slc44a2 |
solute carrier family 44, member 2 |
4513 |
0.11 |
| chr5_118480209_118480360 | 0.95 |
Gm15754 |
predicted gene 15754 |
6683 |
0.21 |
| chr13_52986026_52986367 | 0.94 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
5123 |
0.19 |
| chr14_76149541_76149852 | 0.93 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
38805 |
0.15 |
| chr3_84475488_84475639 | 0.93 |
Fhdc1 |
FH2 domain containing 1 |
3419 |
0.29 |
| chr11_64839925_64840124 | 0.92 |
Gm12292 |
predicted gene 12292 |
3729 |
0.32 |
| chr12_117744512_117744705 | 0.92 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
4013 |
0.25 |
| chr12_110975345_110976350 | 0.92 |
Ankrd9 |
ankyrin repeat domain 9 |
2408 |
0.19 |
| chr17_56756624_56758021 | 0.91 |
Nrtn |
neurturin |
208 |
0.88 |
| chr2_104498055_104498315 | 0.91 |
Hipk3 |
homeodomain interacting protein kinase 3 |
3739 |
0.19 |
| chr8_116501999_116502150 | 0.90 |
Dynlrb2 |
dynein light chain roadblock-type 2 |
2900 |
0.32 |
| chr15_83606896_83607158 | 0.90 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
6712 |
0.15 |
| chr13_75707504_75708793 | 0.89 |
Ell2 |
elongation factor RNA polymerase II 2 |
437 |
0.65 |
| chr11_72429181_72429515 | 0.88 |
Ggt6 |
gamma-glutamyltransferase 6 |
6178 |
0.13 |
| chr16_15948263_15948760 | 0.86 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
60095 |
0.1 |
| chr17_26509441_26509592 | 0.85 |
Dusp1 |
dual specificity phosphatase 1 |
997 |
0.39 |
| chr4_59196198_59196829 | 0.81 |
Ugcg |
UDP-glucose ceramide glucosyltransferase |
6955 |
0.18 |
| chr4_89524955_89525174 | 0.81 |
Gm12608 |
predicted gene 12608 |
80420 |
0.09 |
| chr7_128296590_128296741 | 0.80 |
BC017158 |
cDNA sequence BC017158 |
911 |
0.38 |
| chr7_127800319_127801635 | 0.79 |
Hsd3b7 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
36 |
0.92 |
| chr13_99100172_99101285 | 0.79 |
Gm807 |
predicted gene 807 |
22 |
0.98 |
| chr4_24216144_24216295 | 0.78 |
Gm11892 |
predicted gene 11892 |
21881 |
0.28 |
| chr6_34862527_34862817 | 0.78 |
Tmem140 |
transmembrane protein 140 |
474 |
0.7 |
| chr12_73507259_73507518 | 0.78 |
Gm34016 |
predicted gene, 34016 |
1282 |
0.42 |
| chr18_54907818_54907969 | 0.78 |
Zfp608 |
zinc finger protein 608 |
12331 |
0.2 |
| chr11_85799424_85800600 | 0.77 |
Bcas3 |
breast carcinoma amplified sequence 3 |
1809 |
0.24 |
| chr19_9978713_9979183 | 0.76 |
Fth1 |
ferritin heavy polypeptide 1 |
1650 |
0.22 |
| chr1_154117439_154118596 | 0.76 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
66 |
0.97 |
| chr1_174171241_174171450 | 0.75 |
Spta1 |
spectrin alpha, erythrocytic 1 |
1431 |
0.24 |
| chr11_95104821_95105967 | 0.75 |
Dlx3 |
distal-less homeobox 3 |
14725 |
0.12 |
| chr12_58265531_58265740 | 0.74 |
Clec14a |
C-type lectin domain family 14, member a |
3655 |
0.34 |
| chr15_82406468_82407080 | 0.74 |
Gm27331 |
predicted gene, 27331 |
379 |
0.39 |
| chr4_40781630_40781889 | 0.74 |
Psenen-ps |
presenilin enhancer gamma secretase subunit, pseudogene |
6195 |
0.14 |
| chr5_33216392_33216677 | 0.74 |
Spon2 |
spondin 2, extracellular matrix protein |
574 |
0.72 |
| chr18_10839643_10840307 | 0.73 |
Gm18764 |
predicted gene, 18764 |
19705 |
0.14 |
| chr7_80994626_80994777 | 0.72 |
Gm44967 |
predicted gene 44967 |
18 |
0.93 |
| chr4_133007332_133008511 | 0.72 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
3339 |
0.22 |
| chr17_5091693_5091844 | 0.71 |
Gm15599 |
predicted gene 15599 |
20342 |
0.23 |
| chr13_37346508_37346993 | 0.71 |
Ly86 |
lymphocyte antigen 86 |
1361 |
0.39 |
| chr10_85128093_85128664 | 0.70 |
Mterf2 |
mitochondrial transcription termination factor 2 |
351 |
0.86 |
| chr6_31124166_31124436 | 0.70 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
13381 |
0.11 |
| chr3_87900872_87901055 | 0.70 |
Hdgf |
heparin binding growth factor |
5358 |
0.11 |
| chr17_40806812_40807076 | 0.69 |
Crisp2 |
cysteine-rich secretory protein 2 |
59 |
0.97 |
| chr2_34565012_34565383 | 0.69 |
Gm13408 |
predicted gene 13408 |
198 |
0.95 |
| chr10_81359308_81359697 | 0.69 |
Mfsd12 |
major facilitator superfamily domain containing 12 |
1950 |
0.11 |
| chr16_49839981_49840391 | 0.67 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
15180 |
0.25 |
| chr11_32296600_32297646 | 0.67 |
Hba-a2 |
hemoglobin alpha, adult chain 2 |
495 |
0.66 |
| chr5_123397462_123398184 | 0.67 |
Gm15747 |
predicted gene 15747 |
134 |
0.82 |
| chr7_45126094_45126702 | 0.67 |
Snord35a |
small nucleolar RNA, C/D box 35A |
37 |
0.47 |
| chr3_87767082_87768667 | 0.66 |
Pear1 |
platelet endothelial aggregation receptor 1 |
1061 |
0.43 |
| chr11_32283784_32284776 | 0.65 |
Hba-a1 |
hemoglobin alpha, adult chain 1 |
469 |
0.66 |
| chr14_88123382_88123800 | 0.65 |
Rps3a2 |
ribosomal protein S3A2 |
217 |
0.93 |
| chr2_4560599_4561970 | 0.65 |
Frmd4a |
FERM domain containing 4A |
1509 |
0.43 |
| chr1_181838056_181838455 | 0.65 |
Gm26004 |
predicted gene, 26004 |
2313 |
0.2 |
| chr11_84872602_84872983 | 0.63 |
Ggnbp2 |
gametogenetin binding protein 2 |
1975 |
0.2 |
| chr2_170148791_170149171 | 0.62 |
Zfp217 |
zinc finger protein 217 |
878 |
0.72 |
| chr10_77440841_77441276 | 0.62 |
Gm35920 |
predicted gene, 35920 |
16859 |
0.15 |
| chr19_8714838_8715424 | 0.61 |
Slc3a2 |
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
698 |
0.32 |
| chr1_164405360_164405594 | 0.61 |
Gm37411 |
predicted gene, 37411 |
19236 |
0.14 |
| chr7_71711939_71712090 | 0.61 |
Gm17988 |
predicted gene, 17988 |
5823 |
0.18 |
| chr5_125392659_125393102 | 0.60 |
Ubc |
ubiquitin C |
2678 |
0.17 |
| chrX_162727288_162727912 | 0.60 |
Gm15201 |
predicted gene 15201 |
9013 |
0.14 |
| chr2_143889252_143889441 | 0.60 |
Gm11686 |
predicted gene 11686 |
8169 |
0.14 |
| chr18_84412019_84412170 | 0.60 |
Zfp407 |
zinc finger protein 407 |
326 |
0.92 |
| chr14_65805868_65806583 | 0.60 |
Pbk |
PDZ binding kinase |
7 |
0.98 |
| chr15_12375225_12376436 | 0.59 |
Pdzd2 |
PDZ domain containing 2 |
36550 |
0.14 |
| chr1_33645834_33645985 | 0.59 |
Gm17813 |
predicted gene, 17813 |
11872 |
0.13 |
| chr11_32245145_32245675 | 0.59 |
Nprl3 |
nitrogen permease regulator-like 3 |
4768 |
0.12 |
| chr2_38997767_38998606 | 0.58 |
Gm13496 |
predicted gene 13496 |
72 |
0.57 |
| chr10_19186197_19186616 | 0.58 |
Gm20139 |
predicted gene, 20139 |
19168 |
0.18 |
| chr15_82378536_82379272 | 0.58 |
Cyp2d22 |
cytochrome P450, family 2, subfamily d, polypeptide 22 |
1339 |
0.18 |
| chr6_82770788_82771321 | 0.57 |
Hk2 |
hexokinase 2 |
1934 |
0.25 |
| chr18_34362164_34362756 | 0.57 |
Reep5 |
receptor accessory protein 5 |
10037 |
0.17 |
| chr11_49764573_49765151 | 0.57 |
Gm23813 |
predicted gene, 23813 |
6971 |
0.14 |
| chr9_53609339_53609759 | 0.57 |
Acat1 |
acetyl-Coenzyme A acetyltransferase 1 |
781 |
0.57 |
| chr5_92933209_92933495 | 0.57 |
Shroom3 |
shroom family member 3 |
5285 |
0.23 |
| chr17_47332265_47332516 | 0.56 |
Mrps10 |
mitochondrial ribosomal protein S10 |
36497 |
0.1 |
| chr6_38835149_38835559 | 0.56 |
Hipk2 |
homeodomain interacting protein kinase 2 |
1870 |
0.4 |
| chr12_78205797_78206679 | 0.56 |
Gm6657 |
predicted gene 6657 |
5272 |
0.17 |
| chr6_91116004_91117353 | 0.56 |
Nup210 |
nucleoporin 210 |
118 |
0.95 |
| chr2_25577192_25580600 | 0.55 |
Ajm1 |
apical junction component 1 |
1001 |
0.25 |
| chr16_38370407_38370971 | 0.55 |
Popdc2 |
popeye domain containing 2 |
1263 |
0.34 |
| chr6_146501386_146502829 | 0.54 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
71 |
0.97 |
| chr9_66305145_66305804 | 0.54 |
Dapk2 |
death-associated protein kinase 2 |
37100 |
0.14 |
| chr5_134314919_134315630 | 0.54 |
Gtf2i |
general transcription factor II I |
514 |
0.7 |
| chr7_28865529_28866495 | 0.54 |
Lgals7 |
lectin, galactose binding, soluble 7 |
1557 |
0.2 |
| chrX_154974620_154974827 | 0.54 |
Gm6645 |
predicted gene 6645 |
2 |
0.96 |
| chr5_149643277_149643428 | 0.54 |
Gm20005 |
predicted gene, 20005 |
1742 |
0.28 |
| chr6_134928964_134929556 | 0.54 |
Lockd |
lncRNA downstream of Cdkn1b |
105 |
0.9 |
| chr8_109613033_109613184 | 0.53 |
Pkd1l3 |
polycystic kidney disease 1 like 3 |
1409 |
0.33 |
| chr2_158166874_158167121 | 0.53 |
Tgm2 |
transglutaminase 2, C polypeptide |
20561 |
0.13 |
| chr3_146407041_146407816 | 0.53 |
Ssx2ip |
synovial sarcoma, X 2 interacting protein |
2450 |
0.2 |
| chr9_106198580_106199667 | 0.52 |
Ppm1m |
protein phosphatase 1M |
17 |
0.95 |
| chr17_29093963_29095348 | 0.52 |
1700023B13Rik |
RIKEN cDNA 1700023B13 gene |
194 |
0.82 |
| chr5_137290070_137291279 | 0.52 |
Ache |
acetylcholinesterase |
1427 |
0.17 |
| chr2_39008141_39008933 | 0.52 |
Arpc5l |
actin related protein 2/3 complex, subunit 5-like |
380 |
0.72 |
| chr14_25377750_25379085 | 0.52 |
Gm26660 |
predicted gene, 26660 |
218 |
0.94 |
| chr5_31697163_31697872 | 0.51 |
Rbks |
ribokinase |
110 |
0.58 |
| chr2_119333754_119334576 | 0.51 |
Dll4 |
delta like canonical Notch ligand 4 |
5627 |
0.13 |
| chr11_59960539_59961087 | 0.51 |
Rasd1 |
RAS, dexamethasone-induced 1 |
4131 |
0.14 |
| chr4_130173825_130175545 | 0.50 |
Tinagl1 |
tubulointerstitial nephritis antigen-like 1 |
6 |
0.97 |
| chr4_106798811_106800382 | 0.50 |
Acot11 |
acyl-CoA thioesterase 11 |
194 |
0.93 |
| chr12_44270345_44270549 | 0.50 |
Pnpla8 |
patatin-like phospholipase domain containing 8 |
1280 |
0.31 |
| chr2_128700549_128700956 | 0.50 |
Mertk |
MER proto-oncogene tyrosine kinase |
1796 |
0.3 |
| chr4_154926533_154926891 | 0.50 |
Tnfrsf14 |
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
1365 |
0.31 |
| chr5_129730231_129730850 | 0.49 |
Gm15903 |
predicted gene 15903 |
2368 |
0.17 |
| chr3_148958783_148960002 | 0.48 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
4605 |
0.24 |
| chr16_58498366_58498517 | 0.48 |
St3gal6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
900 |
0.59 |
| chr9_32940723_32941376 | 0.48 |
Gm27162 |
predicted gene 27162 |
12083 |
0.21 |
| chr2_70425556_70425707 | 0.48 |
Rpl9-ps7 |
ribosomal protein L9, pseudogene 7 |
33351 |
0.14 |
| chr19_57361091_57362043 | 0.47 |
Fam160b1 |
family with sequence similarity 160, member B1 |
537 |
0.55 |
| chr8_94035053_94035555 | 0.47 |
Nudt21 |
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
1702 |
0.22 |
| chr4_63557098_63557688 | 0.47 |
Tmem268 |
transmembrane protein 268 |
1388 |
0.3 |
| chr12_47139542_47139693 | 0.47 |
Gm36971 |
predicted gene, 36971 |
25425 |
0.24 |
| chr15_77931320_77931782 | 0.47 |
Txn2 |
thioredoxin 2 |
2545 |
0.21 |
| chr17_35896363_35897367 | 0.46 |
Gm16279 |
predicted gene 16279 |
81 |
0.64 |
| chr19_15941445_15941936 | 0.46 |
Gm3329 |
predicted gene 3329 |
1920 |
0.31 |
| chr16_96192359_96192600 | 0.46 |
Lca5l |
Leber congenital amaurosis 5-like |
208 |
0.92 |
| chr5_31048714_31049304 | 0.45 |
Slc5a6 |
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
85 |
0.68 |
| chr8_84203803_84204350 | 0.45 |
Gm37352 |
predicted gene, 37352 |
3207 |
0.07 |
| chr14_30921218_30921369 | 0.45 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
2256 |
0.18 |
| chr17_57248057_57248976 | 0.45 |
Gpr108 |
G protein-coupled receptor 108 |
70 |
0.92 |
| chr5_113945581_113945982 | 0.45 |
Gm42797 |
predicted gene 42797 |
15647 |
0.11 |
| chr14_101872557_101872708 | 0.44 |
Lmo7 |
LIM domain only 7 |
3162 |
0.35 |
| chr7_25385870_25386460 | 0.44 |
Lipe |
lipase, hormone sensitive |
1239 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.5 | 1.8 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 | 1.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 0.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.6 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.2 | 1.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 0.6 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.2 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.4 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 1.3 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 0.4 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.9 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.2 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.8 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.1 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.3 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.3 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 1.8 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 1.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.5 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0016553 | adenosine to inosine editing(GO:0006382) base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.3 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.0 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.2 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0090156 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.8 | GO:0021675 | nerve development(GO:0021675) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) |
| 0.0 | 0.0 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 1.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 1.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.3 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 1.5 | GO:0030487 | inositol-4,5-bisphosphate 5-phosphatase activity(GO:0030487) |
| 0.1 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0034843 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 2.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0018572 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |