Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
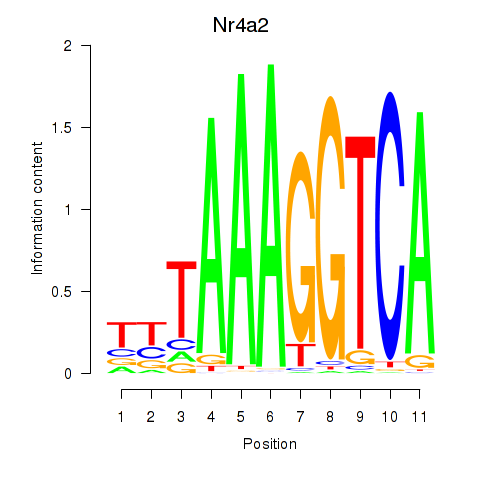
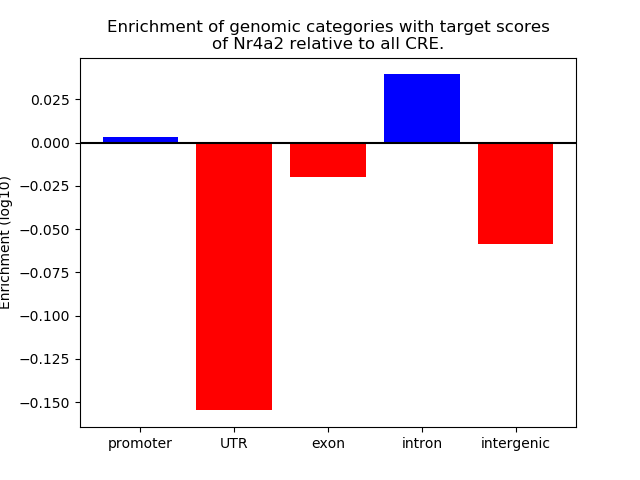
Results for Nr4a2
Z-value: 0.67
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSMUSG00000026826.7 | nuclear receptor subfamily 4, group A, member 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_57108369_57108639 | Nr4a2 | 4568 | 0.231462 | -0.33 | 1.0e-02 | Click! |
| chr2_57108036_57108294 | Nr4a2 | 4907 | 0.227124 | -0.32 | 1.3e-02 | Click! |
| chr2_57103499_57104216 | Nr4a2 | 9215 | 0.204004 | 0.27 | 4.0e-02 | Click! |
| chr2_57113017_57113298 | Nr4a2 | 85 | 0.974064 | -0.24 | 6.0e-02 | Click! |
| chr2_57121724_57121895 | Nr4a2 | 2194 | 0.317606 | 0.24 | 6.1e-02 | Click! |
Activity of the Nr4a2 motif across conditions
Conditions sorted by the z-value of the Nr4a2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
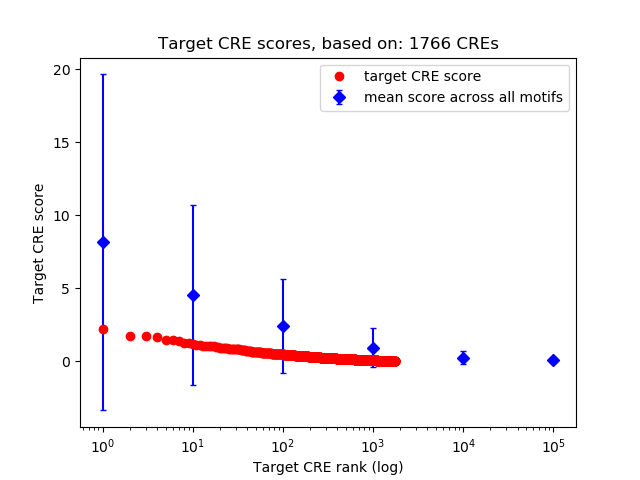
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_32137776_32137927 | 2.22 |
Fosl2 |
fos-like antigen 2 |
1678 |
0.31 |
| chr10_71158552_71158735 | 1.72 |
Bicc1 |
BicC family RNA binding protein 1 |
1047 |
0.39 |
| chr11_90032262_90032532 | 1.70 |
Tmem100 |
transmembrane protein 100 |
2049 |
0.38 |
| chr11_65266805_65267363 | 1.69 |
Myocd |
myocardin |
2770 |
0.29 |
| chr3_133233594_133234317 | 1.48 |
Arhgef38 |
Rho guanine nucleotide exchange factor (GEF) 38 |
934 |
0.64 |
| chr11_5802324_5803834 | 1.44 |
Pgam2 |
phosphoglycerate mutase 2 |
654 |
0.56 |
| chr4_120813489_120813640 | 1.42 |
Nfyc |
nuclear transcription factor-Y gamma |
2148 |
0.21 |
| chr3_5089996_5090891 | 1.27 |
Gm17308 |
predicted gene, 17308 |
167 |
0.97 |
| chr12_35532487_35532918 | 1.26 |
Gm48236 |
predicted gene, 48236 |
1719 |
0.29 |
| chr3_24609075_24609226 | 1.19 |
Gm24704 |
predicted gene, 24704 |
47076 |
0.2 |
| chr13_12337560_12337849 | 1.10 |
Actn2 |
actinin alpha 2 |
3020 |
0.23 |
| chr14_68581840_68582088 | 1.09 |
Adamdec1 |
ADAM-like, decysin 1 |
131 |
0.97 |
| chr6_10612444_10612595 | 1.07 |
Gm2691 |
predicted gene 2691 |
162495 |
0.04 |
| chr1_134802830_134803774 | 1.06 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
549 |
0.61 |
| chr1_151347755_151347925 | 1.05 |
Gm10138 |
predicted gene 10138 |
2609 |
0.2 |
| chr4_132655459_132655610 | 1.04 |
Eya3 |
EYA transcriptional coactivator and phosphatase 3 |
1166 |
0.45 |
| chr6_125322165_125322764 | 1.02 |
Scnn1a |
sodium channel, nonvoltage-gated 1 alpha |
943 |
0.39 |
| chr18_76200378_76201192 | 0.99 |
Gm9028 |
predicted gene 9028 |
23398 |
0.15 |
| chr10_58396489_58397099 | 0.95 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
2406 |
0.3 |
| chr4_140665169_140665889 | 0.92 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
483 |
0.74 |
| chr6_34601698_34602422 | 0.91 |
Cald1 |
caldesmon 1 |
3440 |
0.25 |
| chr4_123235360_123236060 | 0.90 |
Heyl |
hairy/enhancer-of-split related with YRPW motif-like |
1857 |
0.21 |
| chr3_151762523_151763341 | 0.89 |
Ifi44l |
interferon-induced protein 44 like |
40 |
0.98 |
| chr1_66861267_66861452 | 0.89 |
Acadl |
acyl-Coenzyme A dehydrogenase, long-chain |
1918 |
0.18 |
| chr2_76805529_76805788 | 0.86 |
Ttn |
titin |
20884 |
0.22 |
| chr8_57530953_57531157 | 0.86 |
Galnt7 |
polypeptide N-acetylgalactosaminyltransferase 7 |
7398 |
0.1 |
| chr10_121263429_121264335 | 0.86 |
Gm35404 |
predicted gene, 35404 |
20880 |
0.14 |
| chr6_37097381_37097532 | 0.86 |
Dgki |
diacylglycerol kinase, iota |
32946 |
0.23 |
| chrX_117306580_117306731 | 0.85 |
Gm14918 |
predicted gene 14918 |
3853 |
0.35 |
| chr6_44105722_44105873 | 0.83 |
Gm35216 |
predicted gene, 35216 |
22442 |
0.28 |
| chr8_106607096_106607247 | 0.83 |
Cdh1 |
cadherin 1 |
3028 |
0.23 |
| chr19_23143541_23144062 | 0.82 |
Klf9 |
Kruppel-like factor 9 |
157 |
0.95 |
| chr5_35727822_35729089 | 0.81 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
557 |
0.73 |
| chr10_81328518_81330009 | 0.80 |
Tbxa2r |
thromboxane A2 receptor |
532 |
0.47 |
| chr1_125431760_125432298 | 0.78 |
Actr3 |
ARP3 actin-related protein 3 |
2617 |
0.33 |
| chr6_15405115_15405989 | 0.77 |
Gm25470 |
predicted gene, 25470 |
571 |
0.83 |
| chr1_173801485_173801725 | 0.77 |
Ifi203-ps |
interferon activated gene 203, pseudogene |
3335 |
0.17 |
| chr5_61905658_61905809 | 0.76 |
G6pd2 |
glucose-6-phosphate dehydrogenase 2 |
96917 |
0.08 |
| chr19_36114670_36114821 | 0.72 |
Ankrd1 |
ankyrin repeat domain 1 (cardiac muscle) |
5165 |
0.21 |
| chr5_46033018_46033485 | 0.72 |
4930405L22Rik |
RIKEN cDNA 4930405L22 gene |
100842 |
0.07 |
| chr15_99848312_99848463 | 0.71 |
Lima1 |
LIM domain and actin binding 1 |
26284 |
0.07 |
| chr10_126143996_126144329 | 0.71 |
Gm4510 |
predicted gene 4510 |
42161 |
0.18 |
| chrX_164144317_164144468 | 0.70 |
Ace2 |
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
3914 |
0.21 |
| chr5_119846002_119846206 | 0.69 |
Tbx5 |
T-box 5 |
9949 |
0.17 |
| chr14_99772626_99773392 | 0.67 |
Gm22970 |
predicted gene, 22970 |
799 |
0.67 |
| chr3_131334397_131335246 | 0.67 |
Sgms2 |
sphingomyelin synthase 2 |
10112 |
0.15 |
| chr5_36396690_36396841 | 0.67 |
Sorcs2 |
sortilin-related VPS10 domain containing receptor 2 |
1342 |
0.46 |
| chr11_65143082_65143421 | 0.65 |
1700086D15Rik |
RIKEN cDNA 1700086D15 gene |
16639 |
0.13 |
| chr7_134498821_134499325 | 0.63 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
89 |
0.98 |
| chr6_64649585_64650116 | 0.63 |
Grid2 |
glutamate receptor, ionotropic, delta 2 |
13338 |
0.27 |
| chr18_38592430_38592730 | 0.63 |
Spry4 |
sprouty RTK signaling antagonist 4 |
8835 |
0.16 |
| chr4_57571014_57571165 | 0.63 |
Pakap |
paralemmin A kinase anchor protein |
2759 |
0.37 |
| chr5_101381446_101381597 | 0.63 |
Cycs-ps2 |
cytochrome c, pseudogene 2 |
75943 |
0.11 |
| chr4_76453237_76453446 | 0.62 |
Gm42303 |
predicted gene, 42303 |
2110 |
0.34 |
| chr16_97461693_97461844 | 0.61 |
Mx1 |
MX dynamin-like GTPase 1 |
910 |
0.47 |
| chr4_41661471_41661697 | 0.61 |
Cntfr |
ciliary neurotrophic factor receptor |
2450 |
0.15 |
| chr12_63552794_63553211 | 0.61 |
Gm48417 |
predicted gene, 48417 |
18861 |
0.25 |
| chr5_14027948_14028480 | 0.59 |
Gm43519 |
predicted gene 43519 |
320 |
0.88 |
| chr7_112681736_112682136 | 0.59 |
2310014F06Rik |
RIKEN cDNA 2310014F06 gene |
1855 |
0.27 |
| chr16_4520955_4521652 | 0.58 |
Srl |
sarcalumenin |
1760 |
0.3 |
| chr1_57613110_57613752 | 0.58 |
Gm17929 |
predicted gene, 17929 |
49380 |
0.14 |
| chr1_59766087_59766553 | 0.57 |
Bmpr2 |
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
2920 |
0.2 |
| chr14_67229549_67230710 | 0.56 |
Gm20675 |
predicted gene 20675 |
3000 |
0.21 |
| chr12_59129998_59130231 | 0.56 |
Mia2 |
MIA SH3 domain ER export factor 2 |
263 |
0.88 |
| chr4_81566393_81566794 | 0.56 |
Gm11765 |
predicted gene 11765 |
104861 |
0.07 |
| chr17_45689173_45689324 | 0.56 |
Tmem63b |
transmembrane protein 63b |
2343 |
0.17 |
| chr3_89436767_89437632 | 0.55 |
Pbxip1 |
pre B cell leukemia transcription factor interacting protein 1 |
443 |
0.64 |
| chr18_4923350_4923782 | 0.55 |
Svil |
supervillin |
1840 |
0.48 |
| chr13_49608792_49609347 | 0.55 |
Ogn |
osteoglycin |
1023 |
0.44 |
| chr10_80985153_80986200 | 0.55 |
Gng7 |
guanine nucleotide binding protein (G protein), gamma 7 |
12514 |
0.09 |
| chr4_44776724_44776875 | 0.54 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16355 |
0.16 |
| chr5_147723576_147724353 | 0.54 |
Flt1 |
FMS-like tyrosine kinase 1 |
2024 |
0.35 |
| chr10_43830540_43831081 | 0.54 |
4933404K13Rik |
RIKEN cDNA 4933404K13 gene |
4230 |
0.19 |
| chr19_34253411_34255499 | 0.54 |
Acta2 |
actin, alpha 2, smooth muscle, aorta |
225 |
0.92 |
| chr3_79887847_79889192 | 0.53 |
Gm36569 |
predicted gene, 36569 |
1713 |
0.31 |
| chr11_114185237_114185641 | 0.53 |
1700001J04Rik |
RIKEN cDNA 1700001J04 gene |
1235 |
0.46 |
| chr14_103514572_103514723 | 0.53 |
Scel |
sciellin |
1226 |
0.53 |
| chr10_28074317_28074675 | 0.53 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
324 |
0.91 |
| chr8_125102195_125102676 | 0.52 |
Disc1 |
disrupted in schizophrenia 1 |
14448 |
0.19 |
| chr11_96190174_96190661 | 0.52 |
Hoxb13 |
homeobox B13 |
3899 |
0.13 |
| chr16_26369170_26369694 | 0.51 |
Cldn1 |
claudin 1 |
2409 |
0.41 |
| chr3_60408400_60409132 | 0.51 |
Mbnl1 |
muscleblind like splicing factor 1 |
64064 |
0.12 |
| chr11_51291972_51292123 | 0.51 |
Col23a1 |
collagen, type XXIII, alpha 1 |
2127 |
0.26 |
| chr15_85205740_85207341 | 0.50 |
Fbln1 |
fibulin 1 |
535 |
0.78 |
| chr6_36389491_36389642 | 0.50 |
9330158H04Rik |
RIKEN cDNA 9330158H04 gene |
139 |
0.94 |
| chrX_157707743_157707894 | 0.50 |
Smpx |
small muscle protein, X-linked |
5220 |
0.17 |
| chr19_3853794_3854481 | 0.50 |
Gm16066 |
predicted gene 16066 |
1718 |
0.18 |
| chr12_110381274_110381502 | 0.50 |
Gm47195 |
predicted gene, 47195 |
56003 |
0.08 |
| chr6_52005555_52005706 | 0.50 |
Skap2 |
src family associated phosphoprotein 2 |
6885 |
0.17 |
| chr15_42623782_42623933 | 0.49 |
Gm17473 |
predicted gene, 17473 |
52384 |
0.15 |
| chr17_81723281_81723432 | 0.49 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
15021 |
0.27 |
| chr7_68278547_68278698 | 0.49 |
Gm16157 |
predicted gene 16157 |
2028 |
0.23 |
| chr12_73586142_73586320 | 0.49 |
Prkch |
protein kinase C, eta |
1388 |
0.43 |
| chr9_24766942_24767354 | 0.49 |
Tbx20 |
T-box 20 |
2532 |
0.28 |
| chr10_20723991_20725012 | 0.48 |
Pde7b |
phosphodiesterase 7B |
195 |
0.96 |
| chr15_56695618_56696135 | 0.48 |
Has2 |
hyaluronan synthase 2 |
1337 |
0.43 |
| chr2_108131521_108131672 | 0.48 |
Gm23749 |
predicted gene, 23749 |
51554 |
0.18 |
| chr13_23914209_23914456 | 0.48 |
Slc17a4 |
solute carrier family 17 (sodium phosphate), member 4 |
636 |
0.52 |
| chr13_31627499_31628464 | 0.48 |
Gm27516 |
predicted gene, 27516 |
1438 |
0.26 |
| chr6_39474108_39474259 | 0.48 |
Dennd2a |
DENN/MADD domain containing 2A |
3859 |
0.19 |
| chr11_115836836_115837044 | 0.47 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
2617 |
0.15 |
| chr13_51611070_51611510 | 0.47 |
Cks2 |
CDC28 protein kinase regulatory subunit 2 |
33942 |
0.13 |
| chr13_81828595_81828746 | 0.46 |
Cetn3 |
centrin 3 |
36693 |
0.15 |
| chr5_20451812_20452080 | 0.46 |
Magi2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
81554 |
0.1 |
| chr11_36277031_36277877 | 0.46 |
Gm12127 |
predicted gene 12127 |
587 |
0.85 |
| chr2_20521781_20522182 | 0.46 |
Etl4 |
enhancer trap locus 4 |
2153 |
0.43 |
| chr10_88502011_88502741 | 0.45 |
Chpt1 |
choline phosphotransferase 1 |
1377 |
0.37 |
| chr6_112273463_112275203 | 0.45 |
Lmcd1 |
LIM and cysteine-rich domains 1 |
575 |
0.71 |
| chr8_12926230_12928559 | 0.45 |
Mcf2l |
mcf.2 transforming sequence-like |
762 |
0.52 |
| chr10_81195163_81195734 | 0.45 |
Atcayos |
ataxia, cerebellar, Cayman type, opposite strand |
757 |
0.35 |
| chr2_116075452_116076592 | 0.45 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
74 |
0.97 |
| chr11_110968533_110968684 | 0.44 |
Kcnj16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
544 |
0.87 |
| chr17_85375214_85375365 | 0.44 |
Rpl31-ps16 |
ribosomal protein L31, pseudogene 16 |
123344 |
0.05 |
| chr5_24731546_24731845 | 0.44 |
Wdr86 |
WD repeat domain 86 |
968 |
0.47 |
| chr5_66080287_66081072 | 0.44 |
Rbm47 |
RNA binding motif protein 47 |
305 |
0.84 |
| chr19_17834325_17834699 | 0.44 |
Pcsk5 |
proprotein convertase subtilisin/kexin type 5 |
3105 |
0.24 |
| chr3_19306199_19306444 | 0.43 |
Pde7a |
phosphodiesterase 7A |
4582 |
0.29 |
| chr18_77776084_77776500 | 0.43 |
Atp5a1 |
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
2300 |
0.2 |
| chr14_58073809_58074297 | 0.43 |
Fgf9 |
fibroblast growth factor 9 |
1062 |
0.51 |
| chr11_83849571_83850989 | 0.43 |
Hnf1b |
HNF1 homeobox B |
217 |
0.83 |
| chr14_20891990_20892476 | 0.42 |
Gm6128 |
predicted pseudogene 6128 |
35812 |
0.13 |
| chr11_85427959_85428376 | 0.42 |
Bcas3 |
breast carcinoma amplified sequence 3 |
37992 |
0.17 |
| chrX_41063710_41063861 | 0.42 |
Gm14576 |
predicted gene 14576 |
135674 |
0.05 |
| chr13_46422196_46422985 | 0.42 |
Rbm24 |
RNA binding motif protein 24 |
768 |
0.72 |
| chr15_6861862_6862444 | 0.41 |
Osmr |
oncostatin M receptor |
12104 |
0.28 |
| chr1_45500349_45500721 | 0.41 |
Col5a2 |
collagen, type V, alpha 2 |
2747 |
0.22 |
| chr14_60506229_60506951 | 0.41 |
Gm29266 |
predicted gene 29266 |
21382 |
0.2 |
| chr2_138277835_138280637 | 0.40 |
Btbd3 |
BTB (POZ) domain containing 3 |
743 |
0.81 |
| chr4_31877174_31877467 | 0.40 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
86777 |
0.09 |
| chr8_61902711_61903006 | 0.40 |
Palld |
palladin, cytoskeletal associated protein |
168 |
0.95 |
| chr3_101598645_101599448 | 0.40 |
Gm42941 |
predicted gene 42941 |
1916 |
0.31 |
| chr7_104314930_104315471 | 0.40 |
Trim12a |
tripartite motif-containing 12A |
143 |
0.53 |
| chr17_65540372_65540935 | 0.40 |
Tmem232 |
transmembrane protein 232 |
91 |
0.96 |
| chr10_33084458_33084609 | 0.40 |
Trdn |
triadin |
972 |
0.66 |
| chrX_77616014_77616165 | 0.40 |
Gm23121 |
predicted gene, 23121 |
36602 |
0.15 |
| chr13_45692562_45692845 | 0.40 |
Gm47460 |
predicted gene, 47460 |
56869 |
0.15 |
| chr10_89769073_89769465 | 0.40 |
Uhrf1bp1l |
UHRF1 (ICBP90) binding protein 1-like |
17931 |
0.18 |
| chr3_30514092_30514862 | 0.39 |
Mecom |
MDS1 and EVI1 complex locus |
4655 |
0.16 |
| chr1_45765924_45766119 | 0.39 |
Gm8307 |
predicted gene 8307 |
2316 |
0.22 |
| chr11_69419130_69420636 | 0.39 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
6208 |
0.09 |
| chr2_91119045_91119896 | 0.39 |
Mybpc3 |
myosin binding protein C, cardiac |
1326 |
0.3 |
| chr1_135839024_135839175 | 0.39 |
Tnnt2 |
troponin T2, cardiac |
1600 |
0.28 |
| chr5_147563744_147564584 | 0.38 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
24887 |
0.17 |
| chr16_78326082_78326665 | 0.38 |
Cxadr |
coxsackie virus and adenovirus receptor |
1166 |
0.43 |
| chr9_97056372_97056523 | 0.38 |
Gm19325 |
predicted gene, 19325 |
10860 |
0.13 |
| chr11_96326559_96328109 | 0.38 |
Hoxb3 |
homeobox B3 |
728 |
0.39 |
| chr3_27687255_27687406 | 0.38 |
Fndc3b |
fibronectin type III domain containing 3B |
23068 |
0.24 |
| chr6_4506970_4507670 | 0.38 |
Gm43921 |
predicted gene, 43921 |
1233 |
0.34 |
| chr16_23102639_23103369 | 0.38 |
Eif4a2 |
eukaryotic translation initiation factor 4A2 |
4440 |
0.07 |
| chr1_173739285_173739489 | 0.38 |
Ifi207 |
interferon activated gene 207 |
2360 |
0.2 |
| chr3_122566763_122566972 | 0.38 |
Gm43569 |
predicted gene 43569 |
1817 |
0.23 |
| chr3_128192124_128192781 | 0.38 |
D030025E07Rik |
RIKEN cDNA D030025E07 gene |
39101 |
0.18 |
| chr11_108224470_108224622 | 0.37 |
Gm11655 |
predicted gene 11655 |
42696 |
0.17 |
| chr14_121983497_121983648 | 0.37 |
Gpr183 |
G protein-coupled receptor 183 |
18377 |
0.16 |
| chrX_9467805_9468124 | 0.37 |
Cybb |
cytochrome b-245, beta polypeptide |
1305 |
0.48 |
| chr11_96306504_96308444 | 0.37 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
564 |
0.48 |
| chr12_108370973_108372659 | 0.37 |
Eml1 |
echinoderm microtubule associated protein like 1 |
859 |
0.58 |
| chr5_103393811_103394094 | 0.37 |
Ptpn13 |
protein tyrosine phosphatase, non-receptor type 13 |
31240 |
0.14 |
| chr9_44655251_44655818 | 0.37 |
Gm47238 |
predicted gene, 47238 |
4613 |
0.08 |
| chr8_47994470_47995597 | 0.37 |
Wwc2 |
WW, C2 and coiled-coil domain containing 2 |
4109 |
0.22 |
| chr4_15172194_15172551 | 0.37 |
Gm11845 |
predicted gene 11845 |
17980 |
0.21 |
| chrX_169037778_169038469 | 0.37 |
Arhgap6 |
Rho GTPase activating protein 6 |
1512 |
0.52 |
| chr5_77092587_77093501 | 0.37 |
Hopxos |
HOP homeobox, opposite strand |
1488 |
0.28 |
| chr2_4917518_4918021 | 0.36 |
Phyh |
phytanoyl-CoA hydroxylase |
1250 |
0.37 |
| chr10_45288857_45289008 | 0.36 |
Popdc3 |
popeye domain containing 3 |
373 |
0.55 |
| chr6_48705812_48706743 | 0.36 |
Gimap6 |
GTPase, IMAP family member 6 |
41 |
0.93 |
| chr8_57213759_57214585 | 0.36 |
Gm17994 |
predicted gene, 17994 |
77880 |
0.08 |
| chr4_55533366_55533571 | 0.36 |
Klf4 |
Kruppel-like factor 4 (gut) |
1002 |
0.5 |
| chr16_52081920_52082231 | 0.36 |
Gm23513 |
predicted gene, 23513 |
23072 |
0.21 |
| chr15_7574807_7574958 | 0.36 |
Gm49206 |
predicted gene, 49206 |
1360 |
0.56 |
| chr10_91249578_91250191 | 0.36 |
Gm18705 |
predicted gene, 18705 |
9460 |
0.17 |
| chr10_50900495_50901766 | 0.35 |
Sim1 |
single-minded family bHLH transcription factor 1 |
5479 |
0.29 |
| chr17_81737002_81738450 | 0.35 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
651 |
0.81 |
| chr10_78165962_78166113 | 0.35 |
Gatd3a |
glutamine amidotransferase like class 1 domain containing 3A |
1405 |
0.3 |
| chr9_79875436_79876355 | 0.35 |
Gm3211 |
predicted gene 3211 |
37305 |
0.12 |
| chr2_167691999_167692442 | 0.35 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
1039 |
0.35 |
| chr10_107901520_107901895 | 0.35 |
Otogl |
otogelin-like |
10427 |
0.26 |
| chr3_151835993_151836144 | 0.35 |
Ptgfr |
prostaglandin F receptor |
45 |
0.88 |
| chr2_163396823_163398215 | 0.34 |
Jph2 |
junctophilin 2 |
430 |
0.77 |
| chr10_19170810_19170961 | 0.34 |
Gm20139 |
predicted gene, 20139 |
3647 |
0.24 |
| chr3_9831684_9831843 | 0.34 |
Pag1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
1600 |
0.33 |
| chr3_51271701_51271852 | 0.34 |
4930577N17Rik |
RIKEN cDNA 4930577N17 gene |
4983 |
0.13 |
| chr15_51083609_51083844 | 0.34 |
Gm48913 |
predicted gene, 48913 |
186087 |
0.03 |
| chr9_49485451_49486485 | 0.34 |
Ttc12 |
tetratricopeptide repeat domain 12 |
247 |
0.93 |
| chr2_29967054_29967205 | 0.34 |
Sptan1 |
spectrin alpha, non-erythrocytic 1 |
1446 |
0.28 |
| chr9_77044568_77044781 | 0.34 |
Tinag |
tubulointerstitial nephritis antigen |
1075 |
0.46 |
| chr19_46714952_46715103 | 0.34 |
As3mt |
arsenic (+3 oxidation state) methyltransferase |
3055 |
0.19 |
| chr15_80367018_80367304 | 0.33 |
Cacna1i |
calcium channel, voltage-dependent, alpha 1I subunit |
22167 |
0.16 |
| chr1_44217202_44217360 | 0.33 |
Mettl21e |
methyltransferase like 21E |
1632 |
0.34 |
| chr10_80584595_80587584 | 0.33 |
Abhd17a |
abhydrolase domain containing 17A |
535 |
0.53 |
| chr4_92859969_92860120 | 0.33 |
Gm22605 |
predicted gene, 22605 |
6538 |
0.32 |
| chr4_141242265_141243901 | 0.33 |
Arhgef19 |
Rho guanine nucleotide exchange factor (GEF) 19 |
190 |
0.9 |
| chr19_25249376_25249588 | 0.32 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
12507 |
0.24 |
| chr2_76980010_76980504 | 0.32 |
Ttn |
titin |
75 |
0.98 |
| chr17_12760964_12761214 | 0.32 |
Igf2r |
insulin-like growth factor 2 receptor |
8575 |
0.12 |
| chr2_65233164_65233335 | 0.32 |
Cobll1 |
Cobl-like 1 |
2746 |
0.25 |
| chr5_30593992_30594194 | 0.32 |
Gm9899 |
predicted gene 9899 |
5474 |
0.14 |
| chr14_63235525_63236294 | 0.32 |
Gata4 |
GATA binding protein 4 |
9339 |
0.17 |
| chr13_81351343_81352153 | 0.32 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
122 |
0.98 |
| chr10_4080039_4080600 | 0.32 |
Gm25515 |
predicted gene, 25515 |
23994 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.6 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 0.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 0.5 | GO:1900212 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 0.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 0.3 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.4 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.3 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.3 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.2 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.2 | GO:0003180 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 0.2 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.3 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.1 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.2 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) trochlear nerve development(GO:0021558) |
| 0.0 | 0.6 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.0 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.0 | 0.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0034351 | negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.7 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0003177 | pulmonary valve development(GO:0003177) pulmonary valve morphogenesis(GO:0003184) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:2000591 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 1.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.1 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.0 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.0 | GO:0098598 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.0 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.3 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.0 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.0 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.0 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0052041 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.0 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:1904238 | pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.2 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.3 | GO:0044449 | contractile fiber part(GO:0044449) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 0.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.7 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.1 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.7 | GO:0052714 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.0 | GO:0036403 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |