Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
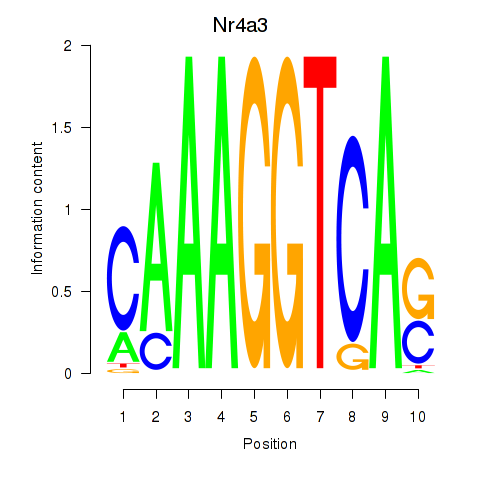
Results for Nr4a3
Z-value: 0.34
Transcription factors associated with Nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a3
|
ENSMUSG00000028341.3 | nuclear receptor subfamily 4, group A, member 3 |
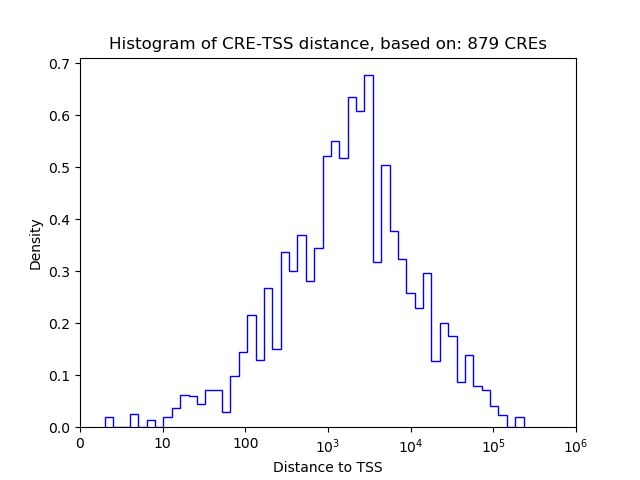
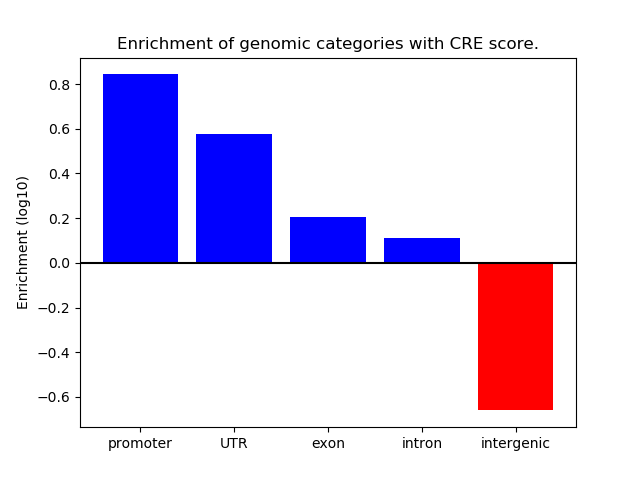
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_48045374_48045835 | Nr4a3 | 373 | 0.892613 | 0.47 | 1.7e-04 | Click! |
| chr4_48046053_48046770 | Nr4a3 | 1180 | 0.547031 | 0.43 | 5.6e-04 | Click! |
| chr4_48046885_48047647 | Nr4a3 | 1848 | 0.400614 | 0.38 | 2.5e-03 | Click! |
| chr4_48047901_48049197 | Nr4a3 | 565 | 0.804093 | 0.35 | 6.3e-03 | Click! |
| chr4_48051161_48052389 | Nr4a3 | 527 | 0.823037 | 0.27 | 3.5e-02 | Click! |
Activity of the Nr4a3 motif across conditions
Conditions sorted by the z-value of the Nr4a3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
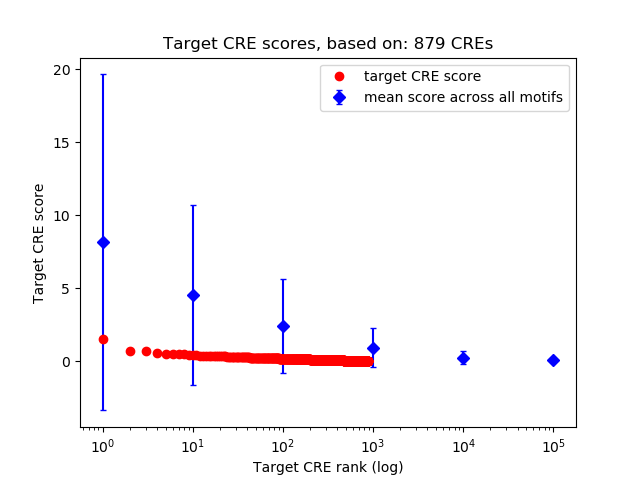
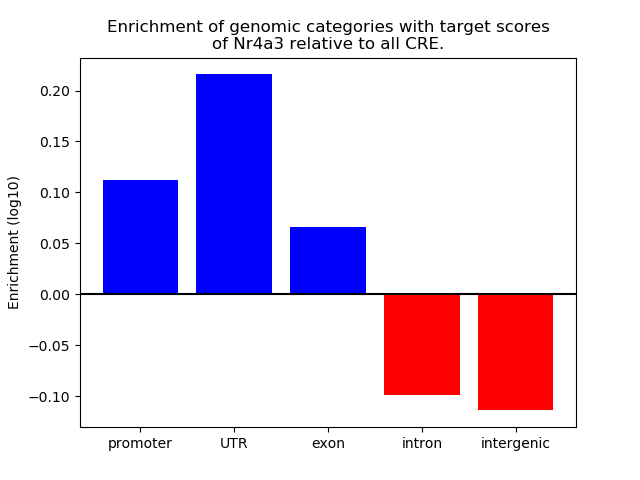
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_170674573_170675954 | 1.51 |
Asmt |
acetylserotonin O-methyltransferase |
2619 |
0.41 |
| chr6_53817061_53817543 | 0.73 |
Gm16499 |
predicted gene 16499 |
1441 |
0.39 |
| chr19_59461344_59461892 | 0.73 |
Emx2 |
empty spiracles homeobox 2 |
1184 |
0.42 |
| chr7_99270744_99271357 | 0.55 |
Map6 |
microtubule-associated protein 6 |
1918 |
0.23 |
| chr13_28948354_28948958 | 0.52 |
Sox4 |
SRY (sex determining region Y)-box 4 |
5057 |
0.24 |
| chr13_53469792_53470715 | 0.50 |
Msx2 |
msh homeobox 2 |
2821 |
0.27 |
| chr12_72234504_72235243 | 0.47 |
Rtn1 |
reticulon 1 |
866 |
0.66 |
| chr3_84303363_84303817 | 0.47 |
Trim2 |
tripartite motif-containing 2 |
1151 |
0.59 |
| chr14_59626952_59628434 | 0.44 |
Shisa2 |
shisa family member 2 |
504 |
0.76 |
| chr5_107498769_107499247 | 0.41 |
Btbd8 |
BTB (POZ) domain containing 8 |
1229 |
0.33 |
| chr2_6883618_6884699 | 0.41 |
Gm13389 |
predicted gene 13389 |
112 |
0.85 |
| chr9_41376046_41377501 | 0.39 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
212 |
0.93 |
| chr10_21882056_21883261 | 0.38 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
289 |
0.9 |
| chr9_91386509_91387874 | 0.37 |
Zic4 |
zinc finger protein of the cerebellum 4 |
4781 |
0.14 |
| chr3_5213504_5214063 | 0.37 |
Gm10748 |
predicted gene 10748 |
945 |
0.56 |
| chr13_31627499_31628464 | 0.36 |
Gm27516 |
predicted gene, 27516 |
1438 |
0.26 |
| chr18_15060980_15061427 | 0.35 |
Kctd1 |
potassium channel tetramerisation domain containing 1 |
686 |
0.74 |
| chr15_101438228_101439215 | 0.35 |
Krt87 |
keratin 87 |
83 |
0.92 |
| chr15_98983254_98984205 | 0.35 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
102 |
0.93 |
| chrX_136957431_136958031 | 0.35 |
Tmsb15b2 |
thymosin beta 15b2 |
294 |
0.8 |
| chr1_62709496_62710533 | 0.34 |
Nrp2 |
neuropilin 2 |
6312 |
0.18 |
| chr3_152106774_152107680 | 0.34 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
195 |
0.8 |
| chr10_14759598_14760682 | 0.33 |
Nmbr |
neuromedin B receptor |
17 |
0.91 |
| chr1_120604474_120605877 | 0.32 |
En1 |
engrailed 1 |
2757 |
0.3 |
| chr11_85838795_85841602 | 0.32 |
Tbx2 |
T-box 2 |
7647 |
0.13 |
| chr2_52860105_52860808 | 0.30 |
Fmnl2 |
formin-like 2 |
2588 |
0.39 |
| chr12_117153278_117156362 | 0.30 |
Gm10421 |
predicted gene 10421 |
3169 |
0.37 |
| chr9_43310784_43311614 | 0.29 |
Trim29 |
tripartite motif-containing 29 |
351 |
0.87 |
| chr19_44496549_44496948 | 0.29 |
Wnt8b |
wingless-type MMTV integration site family, member 8B |
3276 |
0.18 |
| chr3_34641628_34643070 | 0.29 |
Gm42692 |
predicted gene 42692 |
915 |
0.41 |
| chr10_39532030_39532710 | 0.29 |
Fyn |
Fyn proto-oncogene |
900 |
0.6 |
| chr2_32605823_32605974 | 0.28 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
1063 |
0.28 |
| chr18_54493213_54493765 | 0.28 |
4933434P08Rik |
RIKEN cDNA 4933434P08 gene |
3095 |
0.29 |
| chr1_93158156_93158394 | 0.28 |
Mab21l4 |
mab-21-like 4 |
2595 |
0.19 |
| chr2_91930544_91931926 | 0.27 |
Mdk |
midkine |
453 |
0.69 |
| chr2_165137192_165137398 | 0.27 |
Gm14397 |
predicted gene 14397 |
22117 |
0.14 |
| chr15_86028772_86030702 | 0.27 |
Celsr1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
4466 |
0.22 |
| chr8_94995272_94995731 | 0.27 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
160 |
0.93 |
| chr5_134101487_134102203 | 0.27 |
Castor2 |
cytosolic arginine sensor for mTORC1 subunit 2 |
1840 |
0.26 |
| chr9_27303775_27304036 | 0.27 |
Igsf9b |
immunoglobulin superfamily, member 9B |
4677 |
0.22 |
| chr10_91967800_91968283 | 0.27 |
Gm31592 |
predicted gene, 31592 |
79212 |
0.1 |
| chr9_71896189_71896351 | 0.26 |
Tcf12 |
transcription factor 12 |
126 |
0.93 |
| chr3_53043747_53045351 | 0.26 |
Gm42901 |
predicted gene 42901 |
2794 |
0.16 |
| chr4_150730352_150730536 | 0.25 |
Gm16079 |
predicted gene 16079 |
51652 |
0.12 |
| chr15_77303570_77303965 | 0.25 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2838 |
0.22 |
| chr1_167687127_167687278 | 0.25 |
Lmx1a |
LIM homeobox transcription factor 1 alpha |
2035 |
0.44 |
| chr9_91352783_91353195 | 0.25 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
2036 |
0.19 |
| chr16_11068986_11070445 | 0.25 |
Snn |
stannin |
3385 |
0.13 |
| chrX_136976179_136976867 | 0.25 |
Tmsb15l |
thymosin beta 15b like |
346 |
0.41 |
| chr7_29310756_29310907 | 0.24 |
Dpf1 |
D4, zinc and double PHD fingers family 1 |
1225 |
0.31 |
| chr5_111580966_111582197 | 0.24 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
13 |
0.98 |
| chrX_7919510_7921219 | 0.24 |
Pcsk1n |
proprotein convertase subtilisin/kexin type 1 inhibitor |
542 |
0.52 |
| chr11_79662174_79662700 | 0.24 |
Rab11fip4 |
RAB11 family interacting protein 4 (class II) |
1875 |
0.21 |
| chr2_29046202_29046836 | 0.23 |
Cfap77 |
cilia and flagella associated protein 77 |
7159 |
0.17 |
| chr4_154600279_154600535 | 0.23 |
Gm13134 |
predicted gene 13134 |
790 |
0.58 |
| chr1_188149633_188149810 | 0.23 |
Gm38315 |
predicted gene, 38315 |
10986 |
0.27 |
| chr5_103393811_103394094 | 0.23 |
Ptpn13 |
protein tyrosine phosphatase, non-receptor type 13 |
31240 |
0.14 |
| chr5_45638962_45640180 | 0.22 |
Fam184b |
family with sequence similarity 184, member B |
43 |
0.69 |
| chr1_106710395_106710955 | 0.22 |
Bcl2 |
B cell leukemia/lymphoma 2 |
2504 |
0.31 |
| chr9_111440499_111440650 | 0.22 |
Dclk3 |
doublecortin-like kinase 3 |
1493 |
0.42 |
| chr3_156564902_156565177 | 0.22 |
Negr1 |
neuronal growth regulator 1 |
2889 |
0.27 |
| chr11_88071226_88071740 | 0.22 |
Vezf1 |
vascular endothelial zinc finger 1 |
2105 |
0.21 |
| chr17_33962389_33962540 | 0.22 |
Vps52 |
VPS52 GARP complex subunit |
716 |
0.29 |
| chr10_79636527_79636678 | 0.21 |
Gm47163 |
predicted gene, 47163 |
1254 |
0.21 |
| chr2_110949491_110950578 | 0.21 |
Ano3 |
anoctamin 3 |
338 |
0.92 |
| chr1_44199495_44199646 | 0.21 |
Gm15833 |
predicted gene 15833 |
12592 |
0.15 |
| chr16_95673997_95674148 | 0.21 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
28003 |
0.19 |
| chr4_22480645_22481269 | 0.21 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
7409 |
0.17 |
| chr11_21999976_22000574 | 0.21 |
Otx1 |
orthodenticle homeobox 1 |
1340 |
0.53 |
| chr12_3891782_3892977 | 0.21 |
Dnmt3a |
DNA methyltransferase 3A |
635 |
0.69 |
| chr14_98164357_98165375 | 0.21 |
Dach1 |
dachshund family transcription factor 1 |
4677 |
0.28 |
| chr2_25708979_25709130 | 0.21 |
A230005M16Rik |
RIKEN cDNA A230005M16 gene |
662 |
0.44 |
| chr2_181313043_181314281 | 0.21 |
Stmn3 |
stathmin-like 3 |
838 |
0.42 |
| chr6_13835523_13837039 | 0.21 |
Gpr85 |
G protein-coupled receptor 85 |
960 |
0.59 |
| chr7_93085111_93085503 | 0.21 |
Gm9934 |
predicted gene 9934 |
4280 |
0.2 |
| chr10_45808761_45809150 | 0.20 |
D030045P18Rik |
RIKEN cDNA D030045P18 gene |
1251 |
0.49 |
| chr3_87949543_87950464 | 0.20 |
Crabp2 |
cellular retinoic acid binding protein II |
1337 |
0.25 |
| chr19_5606660_5606811 | 0.20 |
Kat5 |
K(lysine) acetyltransferase 5 |
1535 |
0.19 |
| chr15_103522891_103523042 | 0.20 |
Pde1b |
phosphodiesterase 1B, Ca2+-calmodulin dependent |
752 |
0.57 |
| chr6_55010251_55010633 | 0.20 |
Ggct |
gamma-glutamyl cyclotransferase |
17492 |
0.15 |
| chr3_86858235_86858443 | 0.20 |
Gm37025 |
predicted gene, 37025 |
6153 |
0.21 |
| chr5_144547334_144548656 | 0.20 |
Nptx2 |
neuronal pentraxin 2 |
2093 |
0.41 |
| chr11_77936512_77937218 | 0.20 |
Sez6 |
seizure related gene 6 |
5826 |
0.14 |
| chr18_43175442_43176118 | 0.20 |
4930588A03Rik |
RIKEN cDNA 4930588A03 gene |
4506 |
0.2 |
| chr6_127870846_127870997 | 0.20 |
Gm38901 |
predicted gene, 38901 |
6325 |
0.16 |
| chr17_17830971_17831711 | 0.19 |
Spaca6 |
sperm acrosome associated 6 |
103 |
0.78 |
| chr8_25545899_25546050 | 0.19 |
Gm16159 |
predicted gene 16159 |
7586 |
0.11 |
| chr8_47284401_47285223 | 0.19 |
Stox2 |
storkhead box 2 |
4550 |
0.27 |
| chr10_81417616_81418054 | 0.19 |
Mir1191b |
microRNA 1191b |
1538 |
0.15 |
| chr13_46161372_46162144 | 0.19 |
Gm10113 |
predicted gene 10113 |
29288 |
0.21 |
| chr19_15903714_15904400 | 0.19 |
Psat1 |
phosphoserine aminotransferase 1 |
5655 |
0.23 |
| chr12_73043242_73043393 | 0.19 |
Six1 |
sine oculis-related homeobox 1 |
502 |
0.81 |
| chr11_23495316_23495877 | 0.18 |
Ahsa2 |
AHA1, activator of heat shock protein ATPase 2 |
1942 |
0.18 |
| chr1_194622071_194623282 | 0.18 |
Plxna2 |
plexin A2 |
2851 |
0.26 |
| chr2_74691003_74692289 | 0.18 |
Hoxd10 |
homeobox D10 |
278 |
0.7 |
| chr10_80188763_80188923 | 0.18 |
Efna2 |
ephrin A2 |
9361 |
0.08 |
| chr2_13129875_13130250 | 0.18 |
Gm38156 |
predicted gene, 38156 |
11627 |
0.19 |
| chr13_72632318_72633242 | 0.18 |
Irx2 |
Iroquois homeobox 2 |
183 |
0.95 |
| chr2_170734259_170734932 | 0.18 |
Dok5 |
docking protein 5 |
2788 |
0.35 |
| chr8_84178723_84178952 | 0.18 |
Mir181d |
microRNA 181d |
50 |
0.57 |
| chr2_157696327_157696590 | 0.18 |
Gm25407 |
predicted gene, 25407 |
17343 |
0.16 |
| chr5_26210819_26210970 | 0.18 |
Gm7361 |
predicted gene 7361 |
46797 |
0.13 |
| chr7_128474182_128474654 | 0.18 |
Gm32816 |
predicted gene, 32816 |
200 |
0.89 |
| chr11_120293329_120293641 | 0.18 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
3692 |
0.13 |
| chr10_67094914_67095382 | 0.18 |
Jmjd1c |
jumonji domain containing 1C |
977 |
0.49 |
| chr2_33369536_33369687 | 0.18 |
Ralgps1 |
Ral GEF with PH domain and SH3 binding motif 1 |
1818 |
0.3 |
| chr7_126364984_126365135 | 0.17 |
Lat |
linker for activation of T cells |
173 |
0.89 |
| chr11_101084072_101084686 | 0.17 |
Coasy |
Coenzyme A synthase |
484 |
0.59 |
| chr7_45304558_45305606 | 0.17 |
Trpm4 |
transient receptor potential cation channel, subfamily M, member 4 |
122 |
0.9 |
| chr1_159524626_159524870 | 0.17 |
Tnr |
tenascin R |
907 |
0.65 |
| chr5_138606179_138606361 | 0.17 |
Gm24082 |
predicted gene, 24082 |
429 |
0.59 |
| chr16_94141250_94141643 | 0.17 |
Sim2 |
single-minded family bHLH transcription factor 2 |
16265 |
0.12 |
| chr13_52976109_52977372 | 0.17 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
4333 |
0.2 |
| chr6_120716777_120717027 | 0.17 |
Cecr2 |
CECR2, histone acetyl-lysine reader |
17656 |
0.17 |
| chr3_132017605_132018083 | 0.17 |
Gm22421 |
predicted gene, 22421 |
64548 |
0.12 |
| chr18_37755572_37756990 | 0.17 |
Gm26672 |
predicted gene, 26672 |
67 |
0.83 |
| chr2_27353287_27353438 | 0.17 |
Vav2 |
vav 2 oncogene |
8882 |
0.17 |
| chr13_23552480_23553015 | 0.17 |
H1f3 |
H1.3 linker histone, cluster member |
515 |
0.38 |
| chr9_42121573_42122781 | 0.17 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
2120 |
0.33 |
| chr4_24245083_24245586 | 0.17 |
Gm11892 |
predicted gene 11892 |
7234 |
0.32 |
| chr1_118983510_118983661 | 0.17 |
Gli2 |
GLI-Kruppel family member GLI2 |
997 |
0.6 |
| chr2_180343222_180343937 | 0.16 |
Gata5 |
GATA binding protein 5 |
8880 |
0.12 |
| chr17_35909041_35910037 | 0.16 |
Atat1 |
alpha tubulin acetyltransferase 1 |
34 |
0.91 |
| chr6_37592234_37592983 | 0.16 |
Gm7463 |
predicted gene 7463 |
51194 |
0.14 |
| chr17_35244956_35245392 | 0.16 |
Mir6975 |
microRNA 6975 |
1099 |
0.17 |
| chr18_43394505_43395255 | 0.16 |
Dpysl3 |
dihydropyrimidinase-like 3 |
1503 |
0.46 |
| chr19_17834325_17834699 | 0.16 |
Pcsk5 |
proprotein convertase subtilisin/kexin type 5 |
3105 |
0.24 |
| chr12_111694916_111695235 | 0.16 |
Gm36635 |
predicted gene, 36635 |
8007 |
0.1 |
| chr12_118855278_118855595 | 0.16 |
Sp8 |
trans-acting transcription factor 8 |
7850 |
0.22 |
| chr14_52016551_52017265 | 0.16 |
Tmem253 |
transmembrane protein 253 |
11 |
0.93 |
| chr8_114603657_114603808 | 0.16 |
Wwox |
WW domain-containing oxidoreductase |
4704 |
0.23 |
| chr16_33941271_33941422 | 0.16 |
Itgb5 |
integrin beta 5 |
5178 |
0.18 |
| chr3_94480513_94480707 | 0.16 |
Riiad1 |
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
269 |
0.78 |
| chr1_75313516_75313667 | 0.16 |
Dnpep |
aspartyl aminopeptidase |
168 |
0.9 |
| chr2_74732545_74733279 | 0.16 |
Hoxd3 |
homeobox D3 |
1 |
0.92 |
| chr4_106915225_106915605 | 0.15 |
Ssbp3 |
single-stranded DNA binding protein 3 |
3117 |
0.27 |
| chr15_83710142_83710772 | 0.15 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
14469 |
0.21 |
| chr18_60645927_60646078 | 0.15 |
Synpo |
synaptopodin |
2270 |
0.28 |
| chr16_6841576_6842071 | 0.15 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
32601 |
0.26 |
| chr2_25353478_25353940 | 0.15 |
Dpp7 |
dipeptidylpeptidase 7 |
835 |
0.33 |
| chr2_127401705_127401856 | 0.15 |
Gpat2 |
glycerol-3-phosphate acyltransferase 2, mitochondrial |
23419 |
0.12 |
| chr4_139819504_139819950 | 0.15 |
Pax7 |
paired box 7 |
13280 |
0.22 |
| chr19_22692617_22693990 | 0.15 |
Trpm3 |
transient receptor potential cation channel, subfamily M, member 3 |
481 |
0.87 |
| chr19_46152579_46153392 | 0.15 |
Gbf1 |
golgi-specific brefeldin A-resistance factor 1 |
405 |
0.77 |
| chr2_93641010_93641161 | 0.15 |
Alx4 |
aristaless-like homeobox 4 |
1299 |
0.54 |
| chr2_74737192_74739622 | 0.15 |
Hoxd3 |
homeobox D3 |
1212 |
0.2 |
| chr14_50896223_50897440 | 0.15 |
Klhl33 |
kelch-like 33 |
625 |
0.49 |
| chr5_115585775_115585926 | 0.15 |
Gcn1 |
GCN1 activator of EIF2AK4 |
3453 |
0.13 |
| chr13_94912810_94913359 | 0.15 |
Otp |
orthopedia homeobox |
30484 |
0.15 |
| chr3_129194633_129194876 | 0.15 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
5124 |
0.2 |
| chr4_129226645_129228191 | 0.15 |
C77080 |
expressed sequence C77080 |
62 |
0.96 |
| chr2_30137332_30137483 | 0.14 |
Tbc1d13 |
TBC1 domain family, member 13 |
377 |
0.75 |
| chr2_4303011_4303677 | 0.14 |
Gm13187 |
predicted gene 13187 |
2177 |
0.24 |
| chr5_96071564_96071715 | 0.14 |
Cnot6l |
CCR4-NOT transcription complex, subunit 6-like |
15294 |
0.23 |
| chr17_44810813_44811180 | 0.14 |
Runx2 |
runt related transcription factor 2 |
3230 |
0.26 |
| chr16_21205203_21206094 | 0.14 |
Ephb3 |
Eph receptor B3 |
893 |
0.56 |
| chr5_13127277_13127474 | 0.14 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
1782 |
0.32 |
| chr2_30808018_30809041 | 0.14 |
Ntmt1 |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
339 |
0.81 |
| chr16_29054299_29054758 | 0.14 |
Gm8253 |
predicted gene 8253 |
103173 |
0.07 |
| chr17_16483864_16484015 | 0.14 |
Gm4786 |
predicted gene 4786 |
73307 |
0.12 |
| chr12_99229579_99229730 | 0.14 |
Gm19898 |
predicted gene, 19898 |
16017 |
0.18 |
| chr10_13982681_13983672 | 0.14 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
16152 |
0.19 |
| chr11_69911679_69911838 | 0.14 |
Neurl4 |
neuralized E3 ubiquitin protein ligase 4 |
722 |
0.32 |
| chr1_172485277_172486996 | 0.14 |
Igsf9 |
immunoglobulin superfamily, member 9 |
3822 |
0.12 |
| chr13_78174144_78174519 | 0.14 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
2701 |
0.19 |
| chr1_172206683_172207090 | 0.14 |
Pea15a |
phosphoprotein enriched in astrocytes 15A |
82 |
0.94 |
| chr14_67414469_67414620 | 0.14 |
4930438E09Rik |
RIKEN cDNA 4930438E09 gene |
16802 |
0.17 |
| chr6_84134951_84135267 | 0.14 |
Dysf |
dysferlin |
11638 |
0.19 |
| chr10_110454344_110454630 | 0.13 |
Nav3 |
neuron navigator 3 |
1717 |
0.42 |
| chr13_54685334_54685505 | 0.13 |
Rnf44 |
ring finger protein 44 |
1012 |
0.43 |
| chr1_92177475_92177971 | 0.13 |
Hdac4 |
histone deacetylase 4 |
2574 |
0.37 |
| chr1_172204323_172204753 | 0.13 |
Pea15a |
phosphoprotein enriched in astrocytes 15A |
2159 |
0.17 |
| chr19_59452548_59452741 | 0.13 |
Emx2 |
empty spiracles homeobox 2 |
5728 |
0.17 |
| chr1_106629507_106629751 | 0.13 |
Gm37053 |
predicted gene, 37053 |
22759 |
0.19 |
| chr7_25402412_25402872 | 0.13 |
Lipe |
lipase, hormone sensitive |
3932 |
0.11 |
| chr15_100825105_100825748 | 0.13 |
Gm35569 |
predicted gene, 35569 |
6965 |
0.16 |
| chr2_157561445_157562129 | 0.13 |
Gm23134 |
predicted gene, 23134 |
660 |
0.4 |
| chr4_129495840_129495991 | 0.13 |
Fam229a |
family with sequence similarity 229, member A |
4675 |
0.1 |
| chr15_102512807_102512958 | 0.13 |
Map3k12 |
mitogen-activated protein kinase kinase kinase 12 |
2090 |
0.16 |
| chr7_145222634_145222785 | 0.13 |
Smim38 |
small integral membrane protein 38 |
14590 |
0.2 |
| chr7_126276115_126276973 | 0.13 |
Sbk1 |
SH3-binding kinase 1 |
3144 |
0.16 |
| chr13_113918234_113918803 | 0.13 |
Arl15 |
ADP-ribosylation factor-like 15 |
123896 |
0.05 |
| chr7_19634772_19634923 | 0.13 |
Clptm1 |
cleft lip and palate associated transmembrane protein 1 |
1330 |
0.24 |
| chr2_164792559_164792710 | 0.13 |
Snx21 |
sorting nexin family member 21 |
982 |
0.28 |
| chr11_109478129_109478280 | 0.13 |
Slc16a6 |
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
4606 |
0.13 |
| chr12_111412598_111413452 | 0.13 |
Exoc3l4 |
exocyst complex component 3-like 4 |
3992 |
0.14 |
| chr11_120616897_120617872 | 0.13 |
Pcyt2 |
phosphate cytidylyltransferase 2, ethanolamine |
485 |
0.48 |
| chr16_34744940_34746455 | 0.13 |
Mylk |
myosin, light polypeptide kinase |
487 |
0.85 |
| chr13_18948406_18949248 | 0.13 |
Amph |
amphiphysin |
429 |
0.8 |
| chr15_79161202_79161377 | 0.13 |
Sox10 |
SRY (sex determining region Y)-box 10 |
3185 |
0.12 |
| chr3_66972322_66972982 | 0.12 |
Shox2 |
short stature homeobox 2 |
5795 |
0.18 |
| chr17_35907096_35907253 | 0.12 |
Atat1 |
alpha tubulin acetyltransferase 1 |
1787 |
0.11 |
| chr4_134779342_134779493 | 0.12 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
11393 |
0.19 |
| chr15_25407259_25407806 | 0.12 |
Basp1 |
brain abundant, membrane attached signal protein 1 |
6166 |
0.16 |
| chr7_46399032_46399239 | 0.12 |
Kcnc1 |
potassium voltage gated channel, Shaw-related subfamily, member 1 |
1487 |
0.35 |
| chr1_192190419_192191087 | 0.12 |
Kcnh1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
21 |
0.98 |
| chr7_28814945_28816569 | 0.12 |
Hnrnpl |
heterogeneous nuclear ribonucleoprotein L |
651 |
0.47 |
| chr19_46714952_46715103 | 0.12 |
As3mt |
arsenic (+3 oxidation state) methyltransferase |
3055 |
0.19 |
| chr6_137570111_137570964 | 0.12 |
Eps8 |
epidermal growth factor receptor pathway substrate 8 |
222 |
0.95 |
| chr15_37432615_37433119 | 0.12 |
Gm15941 |
predicted gene 15941 |
203 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.1 | 0.2 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.1 | 0.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.0 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:2001201 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0060460 | left lung morphogenesis(GO:0060460) endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.2 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |