Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
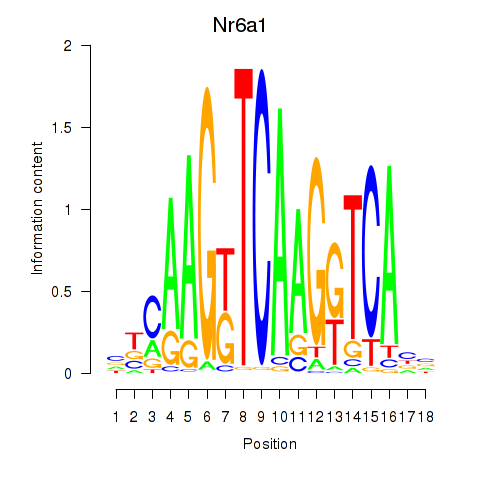
Results for Nr6a1
Z-value: 0.63
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.7 | nuclear receptor subfamily 6, group A, member 1 |
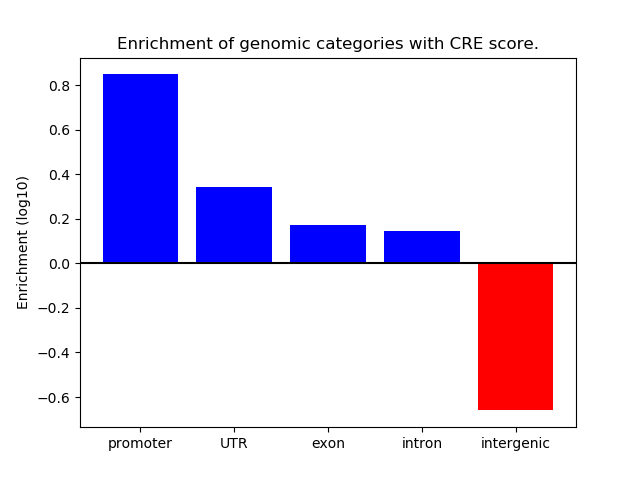
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_38784133_38784675 | Nr6a1 | 65 | 0.954810 | 0.25 | 5.0e-02 | Click! |
| chr2_38924469_38924620 | Nr6a1 | 1673 | 0.230824 | -0.25 | 5.8e-02 | Click! |
| chr2_38927552_38927938 | Nr6a1 | 57 | 0.951716 | 0.23 | 7.6e-02 | Click! |
| chr2_38923092_38923296 | Nr6a1 | 3023 | 0.147837 | -0.23 | 7.8e-02 | Click! |
| chr2_38915140_38915323 | Nr6a1 | 10986 | 0.105994 | -0.23 | 7.8e-02 | Click! |
Activity of the Nr6a1 motif across conditions
Conditions sorted by the z-value of the Nr6a1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
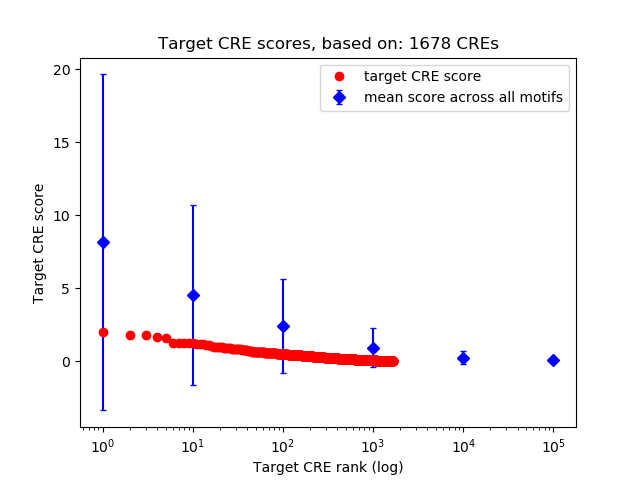
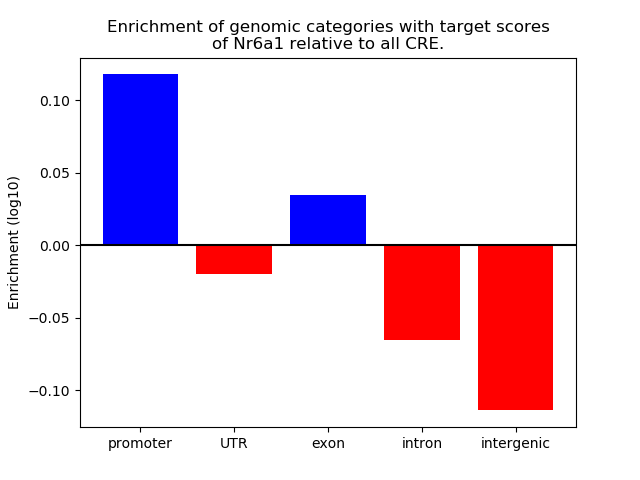
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_163395773_163396078 | 2.01 |
Jph2 |
junctophilin 2 |
2024 |
0.24 |
| chr5_32137776_32137927 | 1.79 |
Fosl2 |
fos-like antigen 2 |
1678 |
0.31 |
| chr3_84189998_84191469 | 1.77 |
Trim2 |
tripartite motif-containing 2 |
210 |
0.94 |
| chr10_97481185_97481829 | 1.65 |
Dcn |
decorin |
860 |
0.67 |
| chr5_102724663_102726037 | 1.57 |
Arhgap24 |
Rho GTPase activating protein 24 |
377 |
0.93 |
| chr19_7558753_7558904 | 1.27 |
Plaat3 |
phospholipase A and acyltransferase 3 |
1343 |
0.39 |
| chr2_71273883_71274736 | 1.26 |
Slc25a12 |
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
8480 |
0.19 |
| chr6_119195676_119196202 | 1.25 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
292 |
0.91 |
| chr11_106381435_106381885 | 1.24 |
Icam2 |
intercellular adhesion molecule 2 |
859 |
0.5 |
| chr9_110765100_110765787 | 1.24 |
Myl3 |
myosin, light polypeptide 3 |
418 |
0.74 |
| chr18_11060814_11061287 | 1.18 |
Gata6 |
GATA binding protein 6 |
2003 |
0.37 |
| chr2_143917101_143917550 | 1.17 |
Dstn |
destrin |
2005 |
0.27 |
| chr5_137610176_137611504 | 1.15 |
Pcolce |
procollagen C-endopeptidase enhancer protein |
138 |
0.86 |
| chr18_80362351_80362669 | 1.10 |
D330025C20Rik |
RIKEN cDNA D330025C20 gene |
271 |
0.79 |
| chr11_119392018_119392570 | 1.10 |
Rnf213 |
ring finger protein 213 |
806 |
0.49 |
| chr18_65393472_65393826 | 1.07 |
Alpk2 |
alpha-kinase 2 |
245 |
0.87 |
| chr15_82378536_82379272 | 0.99 |
Cyp2d22 |
cytochrome P450, family 2, subfamily d, polypeptide 22 |
1339 |
0.18 |
| chr10_68541127_68542141 | 0.99 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
262 |
0.94 |
| chr1_171439752_171440231 | 0.98 |
F11r |
F11 receptor |
2412 |
0.15 |
| chr8_13202008_13202379 | 0.97 |
2810030D12Rik |
RIKEN cDNA 2810030D12 gene |
1373 |
0.23 |
| chr4_32363647_32363850 | 0.96 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
53687 |
0.15 |
| chr3_129330284_129331144 | 0.93 |
Enpep |
glutamyl aminopeptidase |
1550 |
0.35 |
| chr6_6212569_6212720 | 0.92 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
4474 |
0.27 |
| chr3_144759210_144759685 | 0.91 |
Clca3a1 |
chloride channel accessory 3A1 |
1394 |
0.3 |
| chr3_129902773_129902924 | 0.89 |
Casp6 |
caspase 6 |
1347 |
0.38 |
| chr16_92356332_92356564 | 0.88 |
Kcne1 |
potassium voltage-gated channel, Isk-related subfamily, member 1 |
2426 |
0.19 |
| chr2_36216770_36217471 | 0.84 |
Gm13431 |
predicted gene 13431 |
295 |
0.84 |
| chr7_24500255_24500896 | 0.84 |
Cadm4 |
cell adhesion molecule 4 |
614 |
0.51 |
| chr11_117231910_117232833 | 0.83 |
Septin9 |
septin 9 |
86 |
0.97 |
| chr1_78817332_78818790 | 0.83 |
Kcne4 |
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
1134 |
0.48 |
| chr9_74110075_74111065 | 0.83 |
Wdr72 |
WD repeat domain 72 |
166 |
0.97 |
| chr14_63318245_63318396 | 0.82 |
Gm49390 |
predicted gene, 49390 |
575 |
0.72 |
| chr19_38042337_38042611 | 0.82 |
Myof |
myoferlin |
877 |
0.52 |
| chr11_43821050_43821315 | 0.82 |
Adra1b |
adrenergic receptor, alpha 1b |
15150 |
0.23 |
| chrX_106135756_106136481 | 0.77 |
Tlr13 |
toll-like receptor 13 |
7086 |
0.16 |
| chr4_19707395_19707716 | 0.77 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
1438 |
0.45 |
| chr10_81128980_81129986 | 0.75 |
Map2k2 |
mitogen-activated protein kinase kinase 2 |
868 |
0.33 |
| chr12_24892433_24892740 | 0.75 |
Mboat2 |
membrane bound O-acyltransferase domain containing 2 |
60955 |
0.09 |
| chr15_103250315_103251530 | 0.75 |
Nfe2 |
nuclear factor, erythroid derived 2 |
543 |
0.62 |
| chr17_47467845_47469202 | 0.74 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
31147 |
0.09 |
| chr1_184846422_184846573 | 0.74 |
Mtarc2 |
mitochondrial amidoxime reducing component 2 |
46 |
0.97 |
| chr18_46668983_46669540 | 0.72 |
Gm3734 |
predicted gene 3734 |
38454 |
0.1 |
| chr7_4751858_4753020 | 0.71 |
Cox6b2 |
cytochrome c oxidase subunit 6B2 |
89 |
0.92 |
| chr1_179059510_179060167 | 0.70 |
Smyd3 |
SET and MYND domain containing 3 |
70493 |
0.13 |
| chr5_51548617_51548781 | 0.69 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
5172 |
0.21 |
| chr15_73750483_73751713 | 0.67 |
Ptp4a3 |
protein tyrosine phosphatase 4a3 |
2724 |
0.24 |
| chr2_59615026_59615433 | 0.67 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
3154 |
0.32 |
| chr5_134745620_134746234 | 0.66 |
Gm30003 |
predicted gene, 30003 |
1115 |
0.35 |
| chr19_46624649_46624800 | 0.66 |
Wbp1l |
WW domain binding protein 1 like |
1323 |
0.35 |
| chr7_118241830_118242099 | 0.65 |
Smg1 |
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
1385 |
0.3 |
| chr11_111067503_111068134 | 0.65 |
Kcnj2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
1654 |
0.5 |
| chr6_58642449_58642948 | 0.63 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
2116 |
0.36 |
| chr18_84091449_84092719 | 0.63 |
Zadh2 |
zinc binding alcohol dehydrogenase, domain containing 2 |
3413 |
0.19 |
| chr1_65112211_65113305 | 0.63 |
Gm28845 |
predicted gene 28845 |
6800 |
0.1 |
| chr6_52257098_52257896 | 0.63 |
9530018H14Rik |
RIKEN cDNA 9530018H14 gene |
37 |
0.92 |
| chr7_110818849_110819014 | 0.62 |
Rnf141 |
ring finger protein 141 |
2352 |
0.23 |
| chr4_111886523_111886676 | 0.62 |
Slc5a9 |
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
2345 |
0.3 |
| chr10_128738683_128738834 | 0.62 |
Dgka |
diacylglycerol kinase, alpha |
5317 |
0.09 |
| chr19_53673750_53674154 | 0.62 |
Rbm20 |
RNA binding motif protein 20 |
3354 |
0.24 |
| chr17_35821748_35823149 | 0.61 |
Ier3 |
immediate early response 3 |
764 |
0.28 |
| chr8_84711904_84712777 | 0.61 |
Nfix |
nuclear factor I/X |
1728 |
0.2 |
| chr7_128270317_128271410 | 0.60 |
Slc5a2 |
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
289 |
0.79 |
| chr4_57569383_57569972 | 0.60 |
Pakap |
paralemmin A kinase anchor protein |
1347 |
0.55 |
| chr16_35659651_35660734 | 0.60 |
Sema5b |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
757 |
0.67 |
| chr17_81732231_81732916 | 0.60 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
5804 |
0.3 |
| chr12_69574100_69574682 | 0.59 |
Vcpkmt |
valosin containing protein lysine (K) methyltransferase |
8544 |
0.14 |
| chr8_83575796_83577746 | 0.59 |
D830024N08Rik |
RIKEN cDNA D830024N08 gene |
727 |
0.44 |
| chr11_100969289_100970720 | 0.59 |
Cavin1 |
caveolae associated 1 |
547 |
0.68 |
| chr12_110843900_110844184 | 0.59 |
Gm26912 |
predicted gene, 26912 |
1174 |
0.27 |
| chr10_30840937_30841237 | 0.58 |
Hey2 |
hairy/enhancer-of-split related with YRPW motif 2 |
928 |
0.55 |
| chr1_74027276_74028287 | 0.58 |
Tns1 |
tensin 1 |
9352 |
0.23 |
| chr5_114175498_114176701 | 0.58 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
193 |
0.92 |
| chr18_60645396_60645912 | 0.58 |
Synpo |
synaptopodin |
2618 |
0.26 |
| chr9_20868528_20869547 | 0.57 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
240 |
0.82 |
| chr7_134222911_134223711 | 0.57 |
Adam12 |
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
1786 |
0.37 |
| chr5_108674331_108674482 | 0.57 |
Slc26a1 |
solute carrier family 26 (sulfate transporter), member 1 |
347 |
0.78 |
| chr8_80497324_80498362 | 0.56 |
Gypa |
glycophorin A |
4062 |
0.27 |
| chr4_132973841_132975368 | 0.56 |
Fgr |
FGR proto-oncogene, Src family tyrosine kinase |
502 |
0.76 |
| chr5_119137444_119138294 | 0.56 |
1700081H04Rik |
RIKEN cDNA 1700081H04 gene |
29635 |
0.2 |
| chr13_53472002_53473141 | 0.55 |
Msx2 |
msh homeobox 2 |
503 |
0.8 |
| chr8_96577050_96577538 | 0.55 |
1700047G07Rik |
RIKEN cDNA 1700047G07 gene |
64952 |
0.13 |
| chr13_38526290_38526876 | 0.55 |
Gm31834 |
predicted gene, 31834 |
820 |
0.43 |
| chr15_85645713_85647012 | 0.54 |
Gm49539 |
predicted gene, 49539 |
279 |
0.89 |
| chr14_63235525_63236294 | 0.53 |
Gata4 |
GATA binding protein 4 |
9339 |
0.17 |
| chr5_144507108_144507703 | 0.53 |
Nptx2 |
neuronal pentraxin 2 |
38497 |
0.17 |
| chr4_98108504_98108946 | 0.53 |
Gm12691 |
predicted gene 12691 |
37874 |
0.2 |
| chr3_123164422_123164582 | 0.52 |
Gm43287 |
predicted gene 43287 |
30 |
0.97 |
| chr2_131559454_131560024 | 0.52 |
Gm14285 |
predicted gene 14285 |
1875 |
0.29 |
| chr2_127729042_127730365 | 0.52 |
Mall |
mal, T cell differentiation protein-like |
229 |
0.9 |
| chrX_157701658_157702782 | 0.51 |
Smpx |
small muscle protein, X-linked |
378 |
0.83 |
| chr10_105526951_105527102 | 0.51 |
Gm15664 |
predicted gene 15664 |
6566 |
0.13 |
| chr11_96074134_96074285 | 0.51 |
Atp5g1 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
828 |
0.42 |
| chr18_65025085_65025364 | 0.51 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
578 |
0.79 |
| chr3_39460233_39460840 | 0.51 |
Gm9845 |
predicted pseudogene 9845 |
101633 |
0.08 |
| chr2_148441004_148443557 | 0.51 |
Cd93 |
CD93 antigen |
1283 |
0.41 |
| chr15_95805007_95805158 | 0.51 |
Gm49173 |
predicted gene, 49173 |
681 |
0.57 |
| chr11_114072204_114072652 | 0.50 |
Sdk2 |
sidekick cell adhesion molecule 2 |
5382 |
0.24 |
| chr12_110487974_110488308 | 0.50 |
Gm19605 |
predicted gene, 19605 |
1933 |
0.27 |
| chr1_195091807_195092906 | 0.50 |
Cd46 |
CD46 antigen, complement regulatory protein |
107 |
0.94 |
| chr1_74806289_74808675 | 0.50 |
Wnt10a |
wingless-type MMTV integration site family, member 10A |
14119 |
0.11 |
| chr7_143106768_143108439 | 0.50 |
Kcnq1 |
potassium voltage-gated channel, subfamily Q, member 1 |
52 |
0.95 |
| chr10_7473717_7473913 | 0.49 |
Ulbp1 |
UL16 binding protein 1 |
177 |
0.95 |
| chr13_93494772_93495201 | 0.49 |
Jmy |
junction-mediating and regulatory protein |
4751 |
0.2 |
| chr10_98572877_98573494 | 0.48 |
Gm34297 |
predicted gene, 34297 |
35703 |
0.2 |
| chr14_62751561_62751747 | 0.48 |
Ints6 |
integrator complex subunit 6 |
9458 |
0.16 |
| chr13_64248041_64249406 | 0.48 |
1810034E14Rik |
RIKEN cDNA 1810034E14 gene |
5 |
0.55 |
| chr13_50958059_50958704 | 0.48 |
Gm19009 |
predicted gene, 19009 |
98225 |
0.07 |
| chr10_107901520_107901895 | 0.47 |
Otogl |
otogelin-like |
10427 |
0.26 |
| chr9_25483755_25485057 | 0.47 |
Eepd1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
1539 |
0.42 |
| chr19_57417994_57418728 | 0.47 |
Gm50271 |
predicted gene, 50271 |
5133 |
0.19 |
| chr1_162217585_162219370 | 0.47 |
Dnm3os |
dynamin 3, opposite strand |
601 |
0.46 |
| chr18_80193737_80193888 | 0.47 |
Rbfa |
ribosome binding factor A |
1391 |
0.23 |
| chr18_12335527_12335869 | 0.47 |
Lama3 |
laminin, alpha 3 |
1879 |
0.31 |
| chr8_123097655_123097806 | 0.46 |
Spg7 |
SPG7, paraplegin matrix AAA peptidase subunit |
3209 |
0.11 |
| chr10_12812894_12813893 | 0.46 |
Utrn |
utrophin |
156 |
0.96 |
| chr19_60873441_60873948 | 0.46 |
Prdx3 |
peroxiredoxin 3 |
862 |
0.51 |
| chr8_107944624_107945451 | 0.46 |
Zfhx3 |
zinc finger homeobox 3 |
2393 |
0.38 |
| chr5_119838189_119838772 | 0.46 |
Tbx5 |
T-box 5 |
2325 |
0.27 |
| chr3_101367155_101367578 | 0.46 |
Igsf3 |
immunoglobulin superfamily, member 3 |
9717 |
0.15 |
| chr4_132397551_132398511 | 0.45 |
Phactr4 |
phosphatase and actin regulator 4 |
168 |
0.89 |
| chr14_105589341_105590435 | 0.45 |
9330188P03Rik |
RIKEN cDNA 9330188P03 gene |
169 |
0.95 |
| chr10_117840985_117841282 | 0.45 |
Rap1b |
RAS related protein 1b |
4902 |
0.16 |
| chr2_135439627_135440202 | 0.45 |
9630028H03Rik |
RIKEN cDNA 9630028H03 gene |
143382 |
0.04 |
| chr13_38145014_38146012 | 0.45 |
Dsp |
desmoplakin |
5781 |
0.18 |
| chr9_95256675_95257394 | 0.45 |
Slc9a9 |
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
135348 |
0.04 |
| chr14_31211131_31211749 | 0.44 |
Tnnc1 |
troponin C, cardiac/slow skeletal |
3108 |
0.13 |
| chr1_152623256_152624209 | 0.44 |
Rgl1 |
ral guanine nucleotide dissociation stimulator,-like 1 |
1297 |
0.51 |
| chr2_70838589_70839075 | 0.44 |
Tlk1 |
tousled-like kinase 1 |
13104 |
0.18 |
| chr8_108886846_108887288 | 0.44 |
Gm38318 |
predicted gene, 38318 |
19505 |
0.2 |
| chrX_16816296_16817707 | 0.44 |
Maob |
monoamine oxidase B |
274 |
0.95 |
| chr9_57763587_57763764 | 0.44 |
Clk3 |
CDC-like kinase 3 |
633 |
0.65 |
| chr1_133798392_133798998 | 0.43 |
Atp2b4 |
ATPase, Ca++ transporting, plasma membrane 4 |
2341 |
0.23 |
| chr19_24029116_24029267 | 0.43 |
Fam189a2 |
family with sequence similarity 189, member A2 |
1828 |
0.33 |
| chr13_38353572_38354204 | 0.43 |
Bmp6 |
bone morphogenetic protein 6 |
7735 |
0.17 |
| chr4_45529877_45530765 | 0.43 |
Shb |
src homology 2 domain-containing transforming protein B |
9 |
0.97 |
| chr6_34609817_34610100 | 0.43 |
Cald1 |
caldesmon 1 |
11338 |
0.19 |
| chr8_57354784_57355185 | 0.43 |
5033428I22Rik |
RIKEN cDNA 5033428I22 gene |
14184 |
0.13 |
| chr8_40863729_40864362 | 0.42 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
1467 |
0.34 |
| chr1_34129998_34130149 | 0.42 |
Dst |
dystonin |
8823 |
0.17 |
| chr15_77202074_77202631 | 0.42 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
1084 |
0.46 |
| chr11_32294517_32294773 | 0.42 |
Hba-a2 |
hemoglobin alpha, adult chain 2 |
1844 |
0.2 |
| chr17_16889285_16889779 | 0.42 |
Gm7483 |
predicted gene 7483 |
177 |
0.95 |
| chr7_127217459_127218630 | 0.42 |
Septin1 |
septin 1 |
187 |
0.85 |
| chr13_52060340_52060549 | 0.42 |
Gm48190 |
predicted gene, 48190 |
36198 |
0.14 |
| chr11_66985344_66985531 | 0.42 |
Gm12299 |
predicted gene 12299 |
16649 |
0.13 |
| chr7_4516649_4516857 | 0.42 |
Tnnt1 |
troponin T1, skeletal, slow |
371 |
0.69 |
| chr13_24806238_24806432 | 0.42 |
BC005537 |
cDNA sequence BC005537 |
4315 |
0.14 |
| chr19_46510443_46510594 | 0.41 |
Trim8 |
tripartite motif-containing 8 |
4142 |
0.18 |
| chr6_54971822_54971973 | 0.41 |
Nod1 |
nucleotide-binding oligomerization domain containing 1 |
236 |
0.91 |
| chr16_12697234_12697440 | 0.41 |
Gm38619 |
predicted gene, 38619 |
7031 |
0.28 |
| chr2_26339789_26340241 | 0.41 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
567 |
0.56 |
| chr15_59680309_59681106 | 0.41 |
Gm20150 |
predicted gene, 20150 |
8594 |
0.22 |
| chr11_11691275_11691620 | 0.41 |
Gm12000 |
predicted gene 12000 |
4992 |
0.18 |
| chr10_103142909_103143169 | 0.40 |
Lrriq1 |
leucine-rich repeats and IQ motif containing 1 |
3336 |
0.27 |
| chr17_57276011_57276681 | 0.40 |
Vav1 |
vav 1 oncogene |
2754 |
0.17 |
| chr1_187608598_187610131 | 0.40 |
Esrrg |
estrogen-related receptor gamma |
332 |
0.91 |
| chr5_74632969_74633120 | 0.40 |
Lnx1 |
ligand of numb-protein X 1 |
9523 |
0.19 |
| chr2_114048099_114048464 | 0.40 |
Actc1 |
actin, alpha, cardiac muscle 1 |
4606 |
0.18 |
| chr9_59684016_59684167 | 0.40 |
Gramd2 |
GRAM domain containing 2 |
3947 |
0.16 |
| chr11_51648294_51649457 | 0.40 |
N4bp3 |
NEDD4 binding protein 3 |
1945 |
0.2 |
| chr15_101138908_101139059 | 0.40 |
Acvrl1 |
activin A receptor, type II-like 1 |
1383 |
0.29 |
| chr19_6968781_6970375 | 0.39 |
Plcb3 |
phospholipase C, beta 3 |
130 |
0.89 |
| chr19_57272093_57272296 | 0.39 |
4930449E18Rik |
RIKEN cDNA 4930449E18 gene |
1766 |
0.37 |
| chr10_116986532_116987280 | 0.39 |
Best3 |
bestrophin 3 |
592 |
0.61 |
| chr6_52251199_52251449 | 0.39 |
Hoxa11os |
homeobox A11, opposite strand |
2902 |
0.08 |
| chr3_9334054_9335134 | 0.39 |
C030034L19Rik |
RIKEN cDNA C030034L19 gene |
68470 |
0.11 |
| chr17_72921491_72924008 | 0.39 |
Lbh |
limb-bud and heart |
1561 |
0.47 |
| chr11_101771824_101772990 | 0.39 |
Etv4 |
ets variant 4 |
226 |
0.91 |
| chr11_88849197_88849437 | 0.39 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
2186 |
0.23 |
| chr2_25054469_25055450 | 0.39 |
Gm24328 |
predicted gene, 24328 |
216 |
0.68 |
| chr7_108929500_108930178 | 0.38 |
Nlrp10 |
NLR family, pyrin domain containing 10 |
339 |
0.81 |
| chr6_118819427_118819578 | 0.38 |
Gm25905 |
predicted gene, 25905 |
15398 |
0.24 |
| chr8_120275935_120277234 | 0.38 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
48128 |
0.12 |
| chr12_73587917_73588487 | 0.38 |
Prkch |
protein kinase C, eta |
3359 |
0.25 |
| chr14_8408866_8409237 | 0.38 |
Gm8416 |
predicted gene 8416 |
134 |
0.96 |
| chr1_43196103_43196958 | 0.38 |
Fhl2 |
four and a half LIM domains 2 |
231 |
0.93 |
| chr8_34752986_34753476 | 0.38 |
Dusp4 |
dual specificity phosphatase 4 |
54066 |
0.12 |
| chr1_186587900_186588171 | 0.38 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
29354 |
0.18 |
| chr18_53249928_53250416 | 0.38 |
Snx24 |
sorting nexing 24 |
4403 |
0.25 |
| chr15_85670939_85672357 | 0.38 |
Lncppara |
long noncoding RNA near Ppara |
18032 |
0.13 |
| chr13_112460134_112460285 | 0.37 |
Il6st |
interleukin 6 signal transducer |
3861 |
0.19 |
| chr1_23274015_23274590 | 0.37 |
Mir30a |
microRNA 30a |
2033 |
0.21 |
| chr13_51408385_51410292 | 0.37 |
S1pr3 |
sphingosine-1-phosphate receptor 3 |
699 |
0.69 |
| chr6_125320913_125322109 | 0.37 |
Scnn1a |
sodium channel, nonvoltage-gated 1 alpha |
10 |
0.96 |
| chr10_77187782_77188266 | 0.37 |
Col18a1 |
collagen, type XVIII, alpha 1 |
21476 |
0.17 |
| chr1_150992792_150993732 | 0.37 |
Hmcn1 |
hemicentin 1 |
173 |
0.95 |
| chr19_5874269_5874420 | 0.37 |
Frmd8 |
FERM domain containing 8 |
895 |
0.33 |
| chr2_11534808_11535247 | 0.36 |
Gm37975 |
predicted gene, 37975 |
333 |
0.84 |
| chr5_138990246_138990397 | 0.36 |
Pdgfa |
platelet derived growth factor, alpha |
3960 |
0.21 |
| chr9_26804636_26804787 | 0.36 |
Glb1l2 |
galactosidase, beta 1-like 2 |
1013 |
0.54 |
| chr19_5821858_5822796 | 0.36 |
Gm27702 |
predicted gene, 27702 |
2380 |
0.12 |
| chr10_97568252_97568843 | 0.36 |
Lum |
lumican |
3419 |
0.23 |
| chr15_78598536_78598827 | 0.35 |
Cyth4 |
cytohesin 4 |
1532 |
0.27 |
| chr10_40285580_40285776 | 0.35 |
Gm47867 |
predicted gene, 47867 |
3625 |
0.13 |
| chr3_108070155_108070321 | 0.35 |
Ampd2 |
adenosine monophosphate deaminase 2 |
5232 |
0.09 |
| chr17_56263000_56263327 | 0.35 |
Fem1a |
fem 1 homolog a |
6353 |
0.09 |
| chr6_24597837_24598262 | 0.35 |
Lmod2 |
leiomodin 2 (cardiac) |
287 |
0.88 |
| chr1_75360071_75361415 | 0.34 |
Des |
desmin |
414 |
0.72 |
| chr10_80347716_80349512 | 0.34 |
Adamtsl5 |
ADAMTS-like 5 |
202 |
0.82 |
| chr9_21228344_21228495 | 0.34 |
Gm16754 |
predicted gene, 16754 |
3016 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.2 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.5 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.5 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.2 | 0.5 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.5 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.3 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.2 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.1 | 0.4 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.1 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.2 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.1 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.1 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.3 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 0.2 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0032803 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.3 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.6 | GO:0060045 | positive regulation of cardiac muscle cell proliferation(GO:0060045) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 1.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.3 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.7 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.7 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0072125 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.0 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.2 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0072530 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0033275 | actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0071351 | response to interleukin-18(GO:0070673) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.0 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0071371 | cellular response to gonadotropin stimulus(GO:0071371) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.1 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.0 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 3.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.8 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 1.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0034812 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |