Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
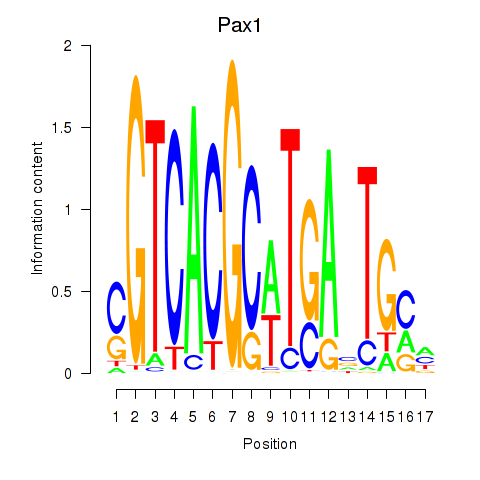
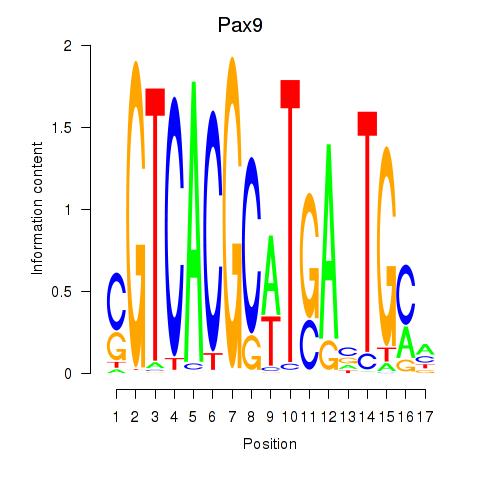
Results for Pax1_Pax9
Z-value: 0.72
Transcription factors associated with Pax1_Pax9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax1
|
ENSMUSG00000037034.9 | paired box 1 |
|
Pax9
|
ENSMUSG00000001497.12 | paired box 9 |
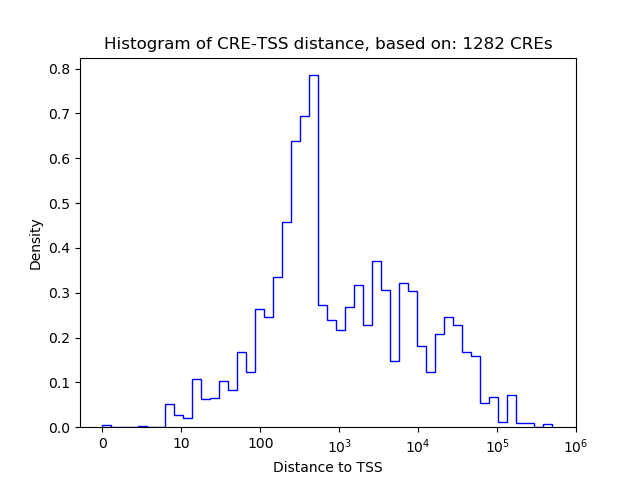
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_147364303_147364454 | Pax1 | 616 | 0.631823 | 0.37 | 4.0e-03 | Click! |
| chr2_147364921_147366397 | Pax1 | 665 | 0.645814 | 0.34 | 8.0e-03 | Click! |
| chr2_147364524_147364675 | Pax1 | 395 | 0.796401 | 0.31 | 1.7e-02 | Click! |
| chr2_147366625_147368623 | Pax1 | 658 | 0.665183 | 0.21 | 1.1e-01 | Click! |
| chr2_147370862_147371013 | Pax1 | 2655 | 0.236025 | 0.20 | 1.3e-01 | Click! |
| chr12_56680898_56682238 | Pax9 | 10199 | 0.137937 | 0.29 | 2.5e-02 | Click! |
| chr12_56704273_56704424 | Pax9 | 8619 | 0.158329 | 0.22 | 9.0e-02 | Click! |
| chr12_56695295_56695580 | Pax9 | 202 | 0.917043 | 0.20 | 1.3e-01 | Click! |
| chr12_56689059_56689356 | Pax9 | 2560 | 0.210570 | -0.19 | 1.4e-01 | Click! |
| chr12_56699459_56700760 | Pax9 | 4380 | 0.177339 | 0.15 | 2.4e-01 | Click! |
Activity of the Pax1_Pax9 motif across conditions
Conditions sorted by the z-value of the Pax1_Pax9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
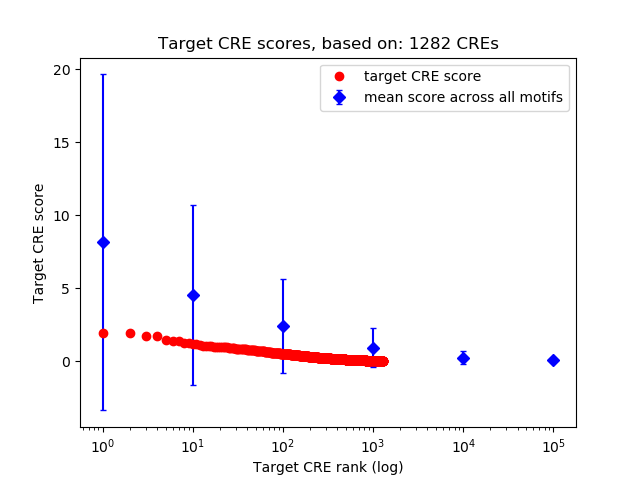
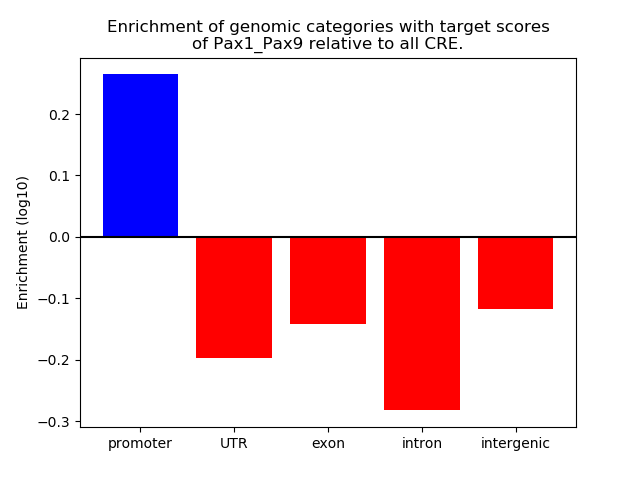
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_38783979_38784442 | 1.93 |
Etv1 |
ets variant 1 |
701 |
0.73 |
| chr3_84334737_84334991 | 1.93 |
Trim2 |
tripartite motif-containing 2 |
27987 |
0.19 |
| chrX_86191680_86192868 | 1.76 |
Nr0b1 |
nuclear receptor subfamily 0, group B, member 1 |
510 |
0.82 |
| chr18_13968574_13969239 | 1.73 |
Zfp521 |
zinc finger protein 521 |
2814 |
0.38 |
| chr2_69129033_69129476 | 1.43 |
Nostrin |
nitric oxide synthase trafficker |
6546 |
0.22 |
| chr3_122026883_122027085 | 1.42 |
Gm42544 |
predicted gene 42544 |
3154 |
0.22 |
| chr1_187439655_187439905 | 1.36 |
Gm37896 |
predicted gene, 37896 |
8097 |
0.26 |
| chr3_117360462_117360799 | 1.29 |
Plppr4 |
phospholipid phosphatase related 4 |
246 |
0.94 |
| chr10_118415370_118415521 | 1.22 |
Ifng |
interferon gamma |
25601 |
0.12 |
| chr3_142168419_142169229 | 1.19 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
389 |
0.89 |
| chr9_108479869_108480980 | 1.17 |
Lamb2 |
laminin, beta 2 |
543 |
0.5 |
| chr18_69343988_69344332 | 1.15 |
Tcf4 |
transcription factor 4 |
14 |
0.99 |
| chr11_43836528_43836860 | 1.08 |
Adra1b |
adrenergic receptor, alpha 1b |
362 |
0.91 |
| chr7_144175485_144176434 | 1.07 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
430 |
0.88 |
| chr17_56605990_56606141 | 1.03 |
Safb |
scaffold attachment factor B |
325 |
0.78 |
| chr17_91085493_91086001 | 1.01 |
Gm47307 |
predicted gene, 47307 |
2659 |
0.21 |
| chr7_48789069_48789872 | 1.01 |
Zdhhc13 |
zinc finger, DHHC domain containing 13 |
438 |
0.8 |
| chr3_138069501_138070764 | 1.00 |
1110002E22Rik |
RIKEN cDNA 1110002E22 gene |
1465 |
0.3 |
| chr11_69235499_69236586 | 0.97 |
Gucy2e |
guanylate cyclase 2e |
980 |
0.38 |
| chr16_18628939_18629866 | 0.97 |
Septin5 |
septin 5 |
302 |
0.83 |
| chr8_69328876_69329533 | 0.96 |
Gm7688 |
predicted gene 7688 |
14 |
0.97 |
| chr18_62930968_62931119 | 0.96 |
Apcdd1 |
adenomatosis polyposis coli down-regulated 1 |
2467 |
0.29 |
| chr2_156196155_156196567 | 0.95 |
Phf20 |
PHD finger protein 20 |
105 |
0.95 |
| chr11_119251895_119252111 | 0.95 |
Ccdc40 |
coiled-coil domain containing 40 |
1550 |
0.27 |
| chr16_75447524_75447796 | 0.94 |
Gm49678 |
predicted gene, 49678 |
367 |
0.91 |
| chr6_21857914_21858140 | 0.92 |
Gm3289 |
predicted gene 3289 |
151 |
0.95 |
| chr3_89322074_89322814 | 0.90 |
Efna3 |
ephrin A3 |
443 |
0.59 |
| chr19_55380884_55381130 | 0.89 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
7 |
0.99 |
| chr9_43744471_43744923 | 0.89 |
Nectin1 |
nectin cell adhesion molecule 1 |
53 |
0.93 |
| chr6_124996427_124996698 | 0.86 |
Pianp |
PILR alpha associated neural protein |
132 |
0.9 |
| chr1_154204657_154205754 | 0.85 |
Zfp648 |
zinc finger protein 648 |
4018 |
0.23 |
| chr6_125690060_125690822 | 0.84 |
Ano2 |
anoctamin 2 |
22 |
0.98 |
| chr11_87442706_87442946 | 0.84 |
Rnu3b1 |
U3B small nuclear RNA 1 |
411 |
0.65 |
| chr13_96180631_96181081 | 0.83 |
Gm29543 |
predicted gene 29543 |
47487 |
0.12 |
| chr5_142904389_142904540 | 0.82 |
Actb |
actin, beta |
810 |
0.55 |
| chr17_75551472_75551866 | 0.82 |
Fam98a |
family with sequence similarity 98, member A |
127 |
0.98 |
| chr4_137993078_137993287 | 0.82 |
Eif4g3 |
eukaryotic translation initiation factor 4 gamma, 3 |
160 |
0.97 |
| chr2_4152602_4153854 | 0.81 |
Frmd4a |
FERM domain containing 4A |
356 |
0.79 |
| chr12_28589414_28590093 | 0.81 |
Allc |
allantoicase |
7230 |
0.16 |
| chr11_76849707_76849858 | 0.80 |
Cpd |
carboxypeptidase D |
2764 |
0.28 |
| chr2_167189983_167190399 | 0.80 |
Kcnb1 |
potassium voltage gated channel, Shab-related subfamily, member 1 |
36 |
0.9 |
| chr5_111725272_111726064 | 0.80 |
Gm26897 |
predicted gene, 26897 |
8256 |
0.19 |
| chr6_89345129_89345500 | 0.79 |
Gm44207 |
predicted gene, 44207 |
174 |
0.93 |
| chr15_89147414_89148183 | 0.77 |
Mapk11 |
mitogen-activated protein kinase 11 |
621 |
0.51 |
| chr6_145048452_145048971 | 0.76 |
Bcat1 |
branched chain aminotransferase 1, cytosolic |
101 |
0.93 |
| chr18_38992539_38992916 | 0.76 |
Arhgap26 |
Rho GTPase activating protein 26 |
418 |
0.79 |
| chr1_25034283_25034476 | 0.76 |
Gm29414 |
predicted gene 29414 |
7147 |
0.21 |
| chr13_49127551_49128199 | 0.76 |
Wnk2 |
WNK lysine deficient protein kinase 2 |
19357 |
0.2 |
| chr6_47044703_47045103 | 0.76 |
Cntnap2 |
contactin associated protein-like 2 |
157 |
0.64 |
| chr7_78884387_78884983 | 0.74 |
Mir7-2 |
microRNA 7-2 |
3592 |
0.14 |
| chr9_35211266_35212010 | 0.74 |
Srpr |
signal recognition particle receptor ('docking protein') |
256 |
0.71 |
| chr15_84104886_84106159 | 0.73 |
Sult4a1 |
sulfotransferase family 4A, member 1 |
87 |
0.93 |
| chr2_84648361_84648948 | 0.73 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
117 |
0.92 |
| chr9_63295779_63296191 | 0.72 |
Map2k5 |
mitogen-activated protein kinase kinase 5 |
7158 |
0.25 |
| chr4_11558571_11558808 | 0.71 |
Rad54b |
RAD54 homolog B (S. cerevisiae) |
233 |
0.92 |
| chr8_95533588_95534657 | 0.70 |
Ccdc113 |
coiled-coil domain containing 113 |
37 |
0.96 |
| chr18_50035394_50035758 | 0.69 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
4547 |
0.23 |
| chr1_37719267_37719774 | 0.69 |
2010300C02Rik |
RIKEN cDNA 2010300C02 gene |
275 |
0.91 |
| chr1_168393023_168393351 | 0.69 |
Pbx1 |
pre B cell leukemia homeobox 1 |
38317 |
0.16 |
| chr4_140630650_140631291 | 0.68 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
13905 |
0.17 |
| chr4_55928799_55929534 | 0.68 |
Gm12519 |
predicted gene 12519 |
64573 |
0.14 |
| chr7_77132082_77132358 | 0.67 |
Gm44879 |
predicted gene 44879 |
166776 |
0.04 |
| chr19_40894409_40894644 | 0.67 |
Mir8092 |
microRNA 8092 |
177 |
0.49 |
| chr2_66100750_66100901 | 0.67 |
Galnt3 |
polypeptide N-acetylgalactosaminyltransferase 3 |
1658 |
0.38 |
| chr6_49821912_49823056 | 0.66 |
Npy |
neuropeptide Y |
226 |
0.95 |
| chr16_52448749_52449233 | 0.66 |
Alcam |
activated leukocyte cell adhesion molecule |
3474 |
0.38 |
| chr8_125235887_125236799 | 0.66 |
Gm16237 |
predicted gene 16237 |
9020 |
0.28 |
| chr2_77775323_77775836 | 0.64 |
Zfp385b |
zinc finger protein 385B |
41237 |
0.17 |
| chr19_8839622_8840720 | 0.62 |
Bscl2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
348 |
0.56 |
| chr2_156477003_156477154 | 0.62 |
Epb41l1 |
erythrocyte membrane protein band 4.1 like 1 |
1544 |
0.28 |
| chr14_32683979_32684180 | 0.62 |
3425401B19Rik |
RIKEN cDNA 3425401B19 gene |
1193 |
0.39 |
| chr18_25067221_25067481 | 0.62 |
Fhod3 |
formin homology 2 domain containing 3 |
57010 |
0.14 |
| chr7_44467574_44467873 | 0.61 |
Josd2 |
Josephin domain containing 2 |
257 |
0.57 |
| chr2_168441196_168441347 | 0.60 |
Gm14234 |
predicted gene 14234 |
22697 |
0.2 |
| chrX_146962362_146963776 | 0.60 |
Htr2c |
5-hydroxytryptamine (serotonin) receptor 2C |
46 |
0.97 |
| chr11_78034745_78034901 | 0.60 |
Dhrs13os |
dehydrogenase/reductase (SDR family) member 13, opposite strand |
2158 |
0.14 |
| chr6_88519047_88519558 | 0.60 |
Gm44430 |
predicted gene, 44430 |
289 |
0.58 |
| chr8_64691882_64692436 | 0.60 |
Cpe |
carboxypeptidase E |
895 |
0.57 |
| chr19_6995553_6996110 | 0.58 |
Nudt22 |
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
206 |
0.56 |
| chr8_66697150_66698637 | 0.58 |
Npy1r |
neuropeptide Y receptor Y1 |
471 |
0.82 |
| chr10_115251625_115252192 | 0.58 |
Tbc1d15 |
TBC1 domain family, member 15 |
441 |
0.82 |
| chr5_129908865_129909017 | 0.58 |
Nupr1l |
nuclear protein transcriptional regulator 1 like |
373 |
0.74 |
| chr5_111256050_111256270 | 0.58 |
Ttc28 |
tetratricopeptide repeat domain 28 |
20845 |
0.18 |
| chr6_122669328_122669812 | 0.57 |
Gm10420 |
predicted gene 10420 |
26431 |
0.1 |
| chr6_126938580_126939554 | 0.56 |
Rad51ap1 |
RAD51 associated protein 1 |
479 |
0.58 |
| chr17_24205669_24205953 | 0.56 |
Tbc1d24 |
TBC1 domain family, member 24 |
249 |
0.8 |
| chr4_143298537_143299593 | 0.55 |
Pdpn |
podoplanin |
398 |
0.83 |
| chr2_91119045_91119896 | 0.55 |
Mybpc3 |
myosin binding protein C, cardiac |
1326 |
0.3 |
| chr15_68931535_68932379 | 0.55 |
Khdrbs3 |
KH domain containing, RNA binding, signal transduction associated 3 |
1889 |
0.38 |
| chr3_123448089_123448441 | 0.55 |
Gm35065 |
predicted gene, 35065 |
818 |
0.45 |
| chr1_79450112_79451070 | 0.55 |
Scg2 |
secretogranin II |
10471 |
0.24 |
| chr16_63863108_63864179 | 0.54 |
Epha3 |
Eph receptor A3 |
230 |
0.96 |
| chr5_24365983_24366141 | 0.54 |
Nos3 |
nitric oxide synthase 3, endothelial cell |
1252 |
0.26 |
| chr6_119669819_119669970 | 0.53 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
47789 |
0.16 |
| chr13_36283360_36283511 | 0.52 |
Gm48766 |
predicted gene, 48766 |
31076 |
0.17 |
| chr9_96889529_96889927 | 0.52 |
Pxylp1 |
2-phosphoxylose phosphatase 1 |
254 |
0.9 |
| chr18_54125378_54126373 | 0.52 |
Gm8594 |
predicted gene 8594 |
94567 |
0.09 |
| chr10_127259299_127259459 | 0.52 |
Kif5a |
kinesin family member 5A |
3701 |
0.09 |
| chr17_64602299_64602450 | 0.51 |
Man2a1 |
mannosidase 2, alpha 1 |
1638 |
0.47 |
| chr5_84281287_84281877 | 0.51 |
Epha5 |
Eph receptor A5 |
80484 |
0.11 |
| chr18_20609699_20610303 | 0.51 |
Dsg2 |
desmoglein 2 |
51755 |
0.1 |
| chr10_29535419_29536227 | 0.51 |
Rspo3 |
R-spondin 3 |
44 |
0.96 |
| chr15_64177808_64178416 | 0.51 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
30379 |
0.19 |
| chr7_80339234_80339763 | 0.50 |
Unc45a |
unc-45 myosin chaperone A |
68 |
0.94 |
| chr9_75541655_75541806 | 0.50 |
Tmod3 |
tropomodulin 3 |
23 |
0.97 |
| chr6_145481872_145482220 | 0.50 |
Gm25373 |
predicted gene, 25373 |
17857 |
0.16 |
| chr4_32916089_32916240 | 0.50 |
Ankrd6 |
ankyrin repeat domain 6 |
7291 |
0.17 |
| chr11_87447972_87448598 | 0.50 |
Rnu3b3 |
U3B small nuclear RNA 3 |
342 |
0.74 |
| chr10_34299043_34301066 | 0.50 |
Tspyl4 |
TSPY-like 4 |
798 |
0.4 |
| chr3_38886705_38888202 | 0.50 |
Fat4 |
FAT atypical cadherin 4 |
513 |
0.62 |
| chr1_175625783_175626003 | 0.49 |
Fh1 |
fumarate hydratase 1 |
258 |
0.91 |
| chr9_30918491_30920140 | 0.48 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
1635 |
0.4 |
| chrX_168779063_168779495 | 0.48 |
Gm15233 |
predicted gene 15233 |
4023 |
0.27 |
| chr11_69412589_69413874 | 0.48 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
444 |
0.64 |
| chr14_54959915_54960463 | 0.48 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
2282 |
0.1 |
| chr16_4267685_4268655 | 0.48 |
Gm5766 |
predicted gene 5766 |
37869 |
0.13 |
| chr8_33711249_33711400 | 0.47 |
Gm24727 |
predicted gene, 24727 |
19523 |
0.12 |
| chr14_67229549_67230710 | 0.47 |
Gm20675 |
predicted gene 20675 |
3000 |
0.21 |
| chr6_83038262_83039190 | 0.47 |
Loxl3 |
lysyl oxidase-like 3 |
326 |
0.68 |
| chr8_106717968_106718245 | 0.47 |
Tango6 |
transport and golgi organization 6 |
34960 |
0.16 |
| chr5_134746440_134747928 | 0.47 |
Eln |
elastin |
62 |
0.61 |
| chr2_69586896_69588383 | 0.46 |
Lrp2 |
low density lipoprotein receptor-related protein 2 |
1574 |
0.42 |
| chr14_101532225_101532447 | 0.46 |
Tbc1d4 |
TBC1 domain family, member 4 |
25010 |
0.19 |
| chr13_71458632_71459335 | 0.45 |
1700112M02Rik |
RIKEN cDNA 1700112M02 gene |
28624 |
0.25 |
| chr19_16872320_16872916 | 0.45 |
Foxb2 |
forkhead box B2 |
1187 |
0.41 |
| chr16_74404583_74404887 | 0.45 |
Robo2 |
roundabout guidance receptor 2 |
6177 |
0.28 |
| chr6_90763313_90763904 | 0.45 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
533 |
0.77 |
| chr13_92530321_92530813 | 0.44 |
Zfyve16 |
zinc finger, FYVE domain containing 16 |
301 |
0.91 |
| chr2_135439627_135440202 | 0.44 |
9630028H03Rik |
RIKEN cDNA 9630028H03 gene |
143382 |
0.04 |
| chr2_137115145_137115830 | 0.44 |
Jag1 |
jagged 1 |
1157 |
0.62 |
| chr17_57769126_57770140 | 0.44 |
Cntnap5c |
contactin associated protein-like 5C |
63 |
0.97 |
| chr2_152636550_152636849 | 0.44 |
Rem1 |
rad and gem related GTP binding protein 1 |
3732 |
0.1 |
| chr7_79466586_79467287 | 0.43 |
Gm10616 |
predicted gene 10616 |
527 |
0.42 |
| chr13_69481908_69482542 | 0.43 |
Gm48676 |
predicted gene, 48676 |
31775 |
0.13 |
| chr4_134102645_134103058 | 0.43 |
Ubxn11 |
UBX domain protein 11 |
232 |
0.87 |
| chr2_166359933_166360116 | 0.43 |
Gm14268 |
predicted gene 14268 |
38133 |
0.16 |
| chr8_54239099_54239565 | 0.42 |
Vegfc |
vascular endothelial growth factor C |
161553 |
0.04 |
| chr12_27334769_27335068 | 0.42 |
Sox11 |
SRY (sex determining region Y)-box 11 |
7656 |
0.29 |
| chrX_94212994_94213232 | 0.42 |
Eif2s3x |
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
251 |
0.91 |
| chr7_130260560_130262022 | 0.42 |
Fgfr2 |
fibroblast growth factor receptor 2 |
566 |
0.84 |
| chr15_23035608_23036941 | 0.42 |
Cdh18 |
cadherin 18 |
181 |
0.97 |
| chr2_26442534_26442685 | 0.41 |
Sec16a |
SEC16 homolog A, endoplasmic reticulum export factor |
2591 |
0.13 |
| chr17_57148993_57149872 | 0.41 |
Cd70 |
CD70 antigen |
345 |
0.79 |
| chr8_11314471_11315722 | 0.41 |
Col4a1 |
collagen, type IV, alpha 1 |
2270 |
0.23 |
| chr2_4154106_4154733 | 0.41 |
Frmd4a |
FERM domain containing 4A |
1547 |
0.26 |
| chr15_98093657_98093849 | 0.41 |
Senp1 |
SUMO1/sentrin specific peptidase 1 |
9 |
0.87 |
| chr7_109438401_109439524 | 0.40 |
Stk33 |
serine/threonine kinase 33 |
36 |
0.83 |
| chr9_77344436_77344707 | 0.40 |
Mlip |
muscular LMNA-interacting protein |
3222 |
0.21 |
| chr16_34744940_34746455 | 0.40 |
Mylk |
myosin, light polypeptide kinase |
487 |
0.85 |
| chr4_124575007_124576219 | 0.40 |
4933407E24Rik |
RIKEN cDNA 4933407E24 gene |
6423 |
0.2 |
| chr12_33027311_33027589 | 0.40 |
Cdhr3 |
cadherin-related family member 3 |
25536 |
0.12 |
| chr4_134245643_134245910 | 0.40 |
Zfp593 |
zinc finger protein 593 |
184 |
0.83 |
| chr17_72925143_72925481 | 0.39 |
Lbh |
limb-bud and heart |
4124 |
0.28 |
| chr2_90474974_90475432 | 0.39 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
3867 |
0.21 |
| chr12_37880304_37881575 | 0.39 |
Dgkb |
diacylglycerol kinase, beta |
223 |
0.96 |
| chr17_45506891_45507081 | 0.39 |
Aars2 |
alanyl-tRNA synthetase 2, mitochondrial |
145 |
0.92 |
| chr2_60722236_60722934 | 0.39 |
Itgb6 |
integrin beta 6 |
58 |
0.98 |
| chr4_66404373_66404645 | 0.38 |
Astn2 |
astrotactin 2 |
26 |
0.99 |
| chr12_89812406_89813090 | 0.38 |
Nrxn3 |
neurexin III |
265 |
0.96 |
| chr9_104062615_104063123 | 0.38 |
Uba5 |
ubiquitin-like modifier activating enzyme 5 |
36 |
0.88 |
| chr6_135011636_135012016 | 0.38 |
Ddx47 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
173 |
0.94 |
| chr11_23067183_23067334 | 0.37 |
Gm12059 |
predicted gene 12059 |
740 |
0.6 |
| chr14_108909943_108910203 | 0.37 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
4085 |
0.37 |
| chr11_78163229_78163523 | 0.37 |
Traf4 |
TNF receptor associated factor 4 |
2140 |
0.11 |
| chr2_59438620_59439113 | 0.37 |
Gm13549 |
predicted gene 13549 |
39085 |
0.13 |
| chr10_117023795_117024543 | 0.37 |
Gm10747 |
predicted gene 10747 |
19577 |
0.11 |
| chr2_19344903_19345364 | 0.36 |
4930447M23Rik |
RIKEN cDNA 4930447M23 gene |
196 |
0.94 |
| chr12_85178232_85178383 | 0.36 |
Pgf |
placental growth factor |
517 |
0.66 |
| chr8_31150331_31150537 | 0.36 |
Tti2 |
TELO2 interacting protein 2 |
94 |
0.85 |
| chr1_153652756_153654124 | 0.36 |
Rgs8 |
regulator of G-protein signaling 8 |
415 |
0.8 |
| chr6_143100273_143100480 | 0.36 |
C2cd5 |
C2 calcium-dependent domain containing 5 |
235 |
0.92 |
| chr8_22506948_22507557 | 0.36 |
Slc20a2 |
solute carrier family 20, member 2 |
1026 |
0.46 |
| chrX_161907356_161907507 | 0.36 |
Gm15202 |
predicted gene 15202 |
792 |
0.75 |
| chr8_121382840_121383267 | 0.35 |
Gm26815 |
predicted gene, 26815 |
26217 |
0.16 |
| chr9_7176368_7177410 | 0.35 |
Dync2h1 |
dynein cytoplasmic 2 heavy chain 1 |
68 |
0.98 |
| chr5_92698277_92699272 | 0.35 |
Shroom3 |
shroom family member 3 |
15149 |
0.16 |
| chr15_96271638_96271789 | 0.35 |
2610037D02Rik |
RIKEN cDNA 2610037D02 gene |
11895 |
0.18 |
| chr11_43528323_43528679 | 0.35 |
Ccnjl |
cyclin J-like |
283 |
0.89 |
| chr17_35867106_35867524 | 0.35 |
Ppp1r18os |
protein phosphatase 1, regulatory subunit 18, opposite strand |
429 |
0.42 |
| chr19_37877611_37878244 | 0.34 |
Gm4757 |
predicted gene 4757 |
7657 |
0.21 |
| chr18_35834043_35834925 | 0.34 |
Gm29417 |
predicted gene 29417 |
3391 |
0.13 |
| chr5_90902965_90904167 | 0.34 |
Cxcl2 |
chemokine (C-X-C motif) ligand 2 |
305 |
0.84 |
| chr18_47365019_47365190 | 0.34 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
3164 |
0.28 |
| chr13_12565430_12565777 | 0.33 |
Ero1lb |
ERO1-like beta (S. cerevisiae) |
271 |
0.88 |
| chr8_56932747_56932927 | 0.33 |
Gm7328 |
predicted gene 7328 |
19992 |
0.23 |
| chr19_7398904_7399138 | 0.33 |
Rab11b-ps2 |
RAB11B, member RAS oncogene family, pseudogene 2 |
56 |
0.95 |
| chr6_85332444_85333044 | 0.33 |
Sfxn5 |
sideroflexin 5 |
6 |
0.97 |
| chr3_106730147_106730298 | 0.33 |
Lrif1 |
ligand dependent nuclear receptor interacting factor 1 |
2692 |
0.25 |
| chr2_73213682_73214373 | 0.33 |
Ola1 |
Obg-like ATPase 1 |
306 |
0.89 |
| chr9_96537234_96537629 | 0.33 |
Gm8495 |
predicted gene 8495 |
270 |
0.86 |
| chr7_141069915_141071420 | 0.33 |
B4galnt4 |
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
65 |
0.93 |
| chr5_135078421_135078620 | 0.33 |
Vps37d |
vacuolar protein sorting 37D |
254 |
0.82 |
| chr10_76468272_76468799 | 0.33 |
Ybey |
ybeY metallopeptidase |
328 |
0.51 |
| chr4_128428742_128429295 | 0.33 |
Csmd2 |
CUB and Sushi multiple domains 2 |
48136 |
0.16 |
| chr18_10706006_10706729 | 0.32 |
Abhd3 |
abhydrolase domain containing 3 |
339 |
0.59 |
| chr4_124570892_124571307 | 0.32 |
4933407E24Rik |
RIKEN cDNA 4933407E24 gene |
1909 |
0.33 |
| chr1_82586379_82587667 | 0.32 |
Col4a3 |
collagen, type IV, alpha 3 |
47 |
0.67 |
| chr18_5333704_5334306 | 0.32 |
Zfp438 |
zinc finger protein 438 |
408 |
0.86 |
| chr6_85373595_85374585 | 0.32 |
Rab11fip5 |
RAB11 family interacting protein 5 (class I) |
474 |
0.76 |
| chr12_46814495_46815083 | 0.31 |
Nova1 |
NOVA alternative splicing regulator 1 |
2171 |
0.31 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.2 | 1.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 1.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.6 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.2 | 0.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 | 0.7 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.2 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.7 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.8 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 1.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.9 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.4 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.2 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.1 | 0.5 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.3 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 1.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.1 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.2 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.1 | 0.2 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:0035482 | gastric motility(GO:0035482) |
| 0.1 | 0.3 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.1 | 0.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.5 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0014900 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.6 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.3 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.0 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0090128 | regulation of synapse maturation(GO:0090128) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:0042851 | alanine metabolic process(GO:0006522) pyruvate family amino acid metabolic process(GO:0009078) L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.0 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0070922 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) small RNA loading onto RISC(GO:0070922) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.0 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 1.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.5 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 1.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 1.3 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.3 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.1 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0034920 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 1.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.2 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0016232 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |