Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
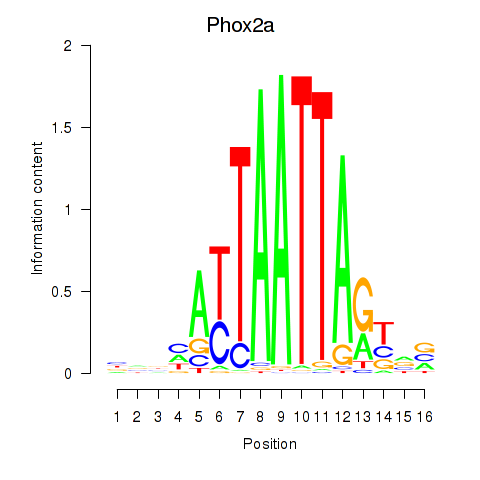
Results for Phox2a
Z-value: 0.44
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.9 | paired-like homeobox 2a |
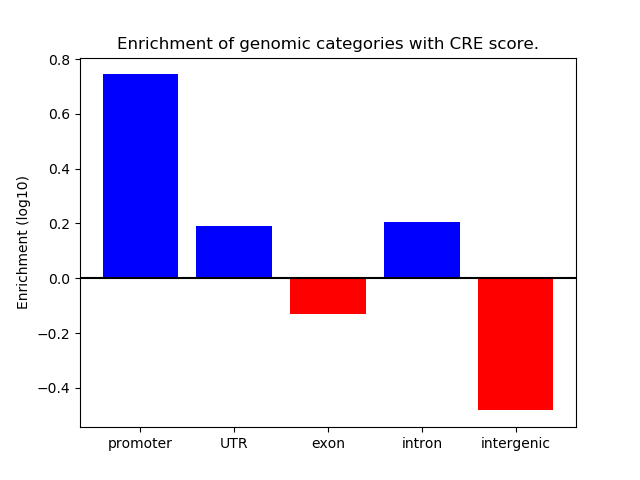
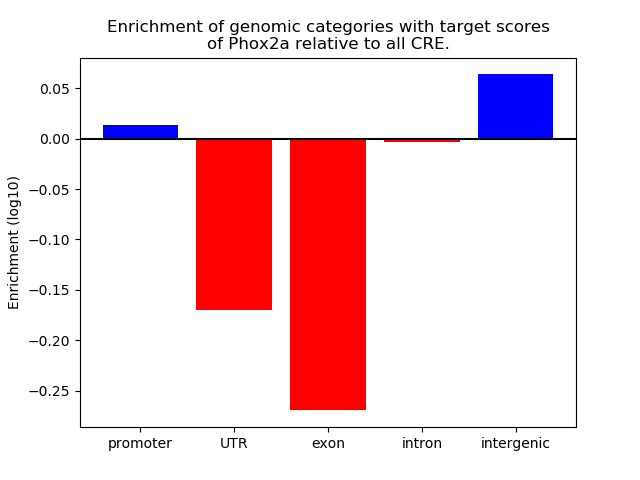
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_101817270_101817487 | Phox2a | 935 | 0.370259 | -0.12 | 3.6e-01 | Click! |
| chr7_101816943_101817222 | Phox2a | 1231 | 0.277841 | -0.11 | 4.1e-01 | Click! |
| chr7_101819111_101819732 | Phox2a | 1108 | 0.305302 | -0.11 | 4.2e-01 | Click! |
| chr7_101819812_101819963 | Phox2a | 859 | 0.396288 | 0.07 | 5.9e-01 | Click! |
| chr7_101820668_101822582 | Phox2a | 879 | 0.377239 | 0.06 | 6.5e-01 | Click! |
Activity of the Phox2a motif across conditions
Conditions sorted by the z-value of the Phox2a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
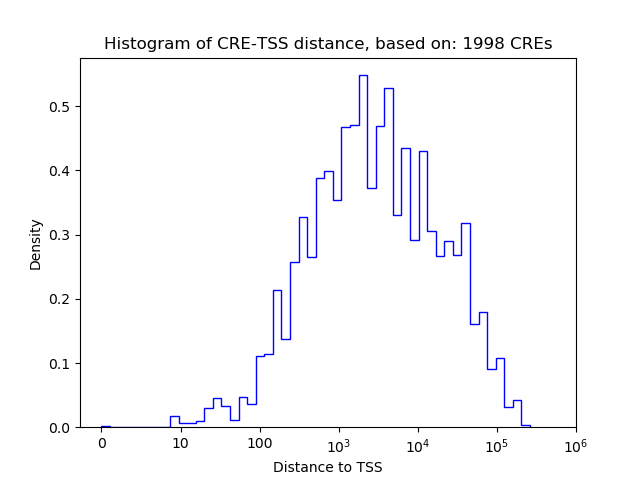
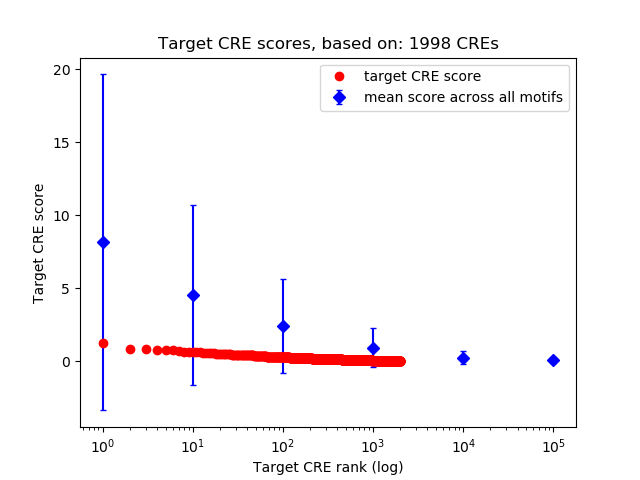
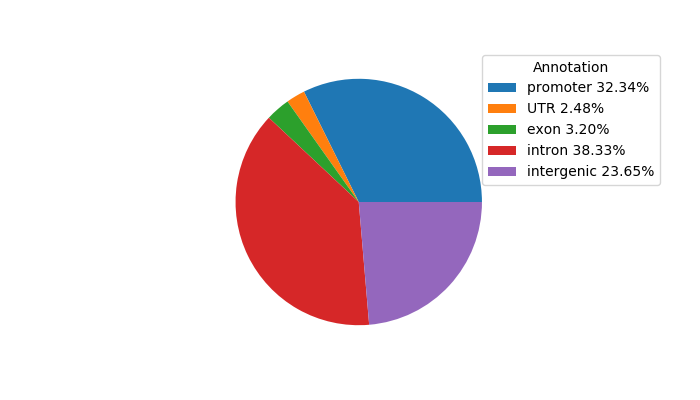
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_63249784_63250324 | 1.23 |
Gata4 |
GATA binding protein 4 |
4783 |
0.2 |
| chr13_96986050_96986201 | 0.87 |
Gm48609 |
predicted gene, 48609 |
29174 |
0.13 |
| chr13_73262153_73264451 | 0.86 |
Irx4 |
Iroquois homeobox 4 |
2805 |
0.22 |
| chr14_111166555_111166800 | 0.79 |
Gm22802 |
predicted gene, 22802 |
115369 |
0.07 |
| chr10_33083500_33083959 | 0.76 |
Trdn |
triadin |
168 |
0.97 |
| chr2_155611238_155612364 | 0.75 |
Myh7b |
myosin, heavy chain 7B, cardiac muscle, beta |
589 |
0.53 |
| chr13_12132755_12132906 | 0.69 |
Ryr2 |
ryanodine receptor 2, cardiac |
25885 |
0.13 |
| chr5_138608490_138608879 | 0.65 |
Zfp68 |
zinc finger protein 68 |
1627 |
0.2 |
| chr7_103809605_103809899 | 0.65 |
Hbb-bt |
hemoglobin, beta adult t chain |
4244 |
0.07 |
| chr17_71290536_71290920 | 0.64 |
Emilin2 |
elastin microfibril interfacer 2 |
6519 |
0.15 |
| chr2_154631717_154631891 | 0.64 |
Gm14198 |
predicted gene 14198 |
832 |
0.46 |
| chr14_32684282_32684433 | 0.63 |
3425401B19Rik |
RIKEN cDNA 3425401B19 gene |
915 |
0.46 |
| chr16_96205810_96206739 | 0.60 |
Sh3bgr |
SH3-binding domain glutamic acid-rich protein |
369 |
0.83 |
| chr17_85485071_85487191 | 0.57 |
Rpl31-ps16 |
ribosomal protein L31, pseudogene 16 |
12502 |
0.23 |
| chr7_136275874_136276030 | 0.55 |
C030029H02Rik |
RIKEN cDNA C030029H02 gene |
7618 |
0.21 |
| chr10_40135263_40135524 | 0.54 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
6861 |
0.14 |
| chr3_134236641_134237783 | 0.54 |
Cxxc4 |
CXXC finger 4 |
392 |
0.78 |
| chr13_7239804_7239955 | 0.51 |
Gm40663 |
predicted gene, 40663 |
63954 |
0.12 |
| chr16_20594716_20594867 | 0.50 |
Vwa5b2 |
von Willebrand factor A domain containing 5B2 |
3635 |
0.07 |
| chr19_53794096_53794765 | 0.50 |
Rbm20 |
RNA binding motif protein 20 |
1122 |
0.48 |
| chr19_28037121_28037467 | 0.49 |
Gm5517 |
predicted gene 5517 |
3830 |
0.23 |
| chr5_138609015_138609166 | 0.49 |
Gm15497 |
predicted gene 15497 |
1649 |
0.2 |
| chrX_142684293_142684706 | 0.48 |
Tmem164 |
transmembrane protein 164 |
1611 |
0.4 |
| chr5_64807638_64809344 | 0.48 |
Klf3 |
Kruppel-like factor 3 (basic) |
3848 |
0.17 |
| chr4_149990023_149990174 | 0.47 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
11167 |
0.15 |
| chr4_123563270_123563441 | 0.47 |
Macf1 |
microtubule-actin crosslinking factor 1 |
1339 |
0.45 |
| chr4_84637359_84638298 | 0.46 |
Bnc2 |
basonuclin 2 |
37168 |
0.19 |
| chr3_149088208_149089249 | 0.46 |
Gm25127 |
predicted gene, 25127 |
59796 |
0.12 |
| chr4_137863933_137864084 | 0.46 |
Ece1 |
endothelin converting enzyme 1 |
1711 |
0.38 |
| chr11_117059965_117061040 | 0.46 |
Gm11731 |
predicted gene 11731 |
2077 |
0.18 |
| chr4_116073957_116074118 | 0.44 |
Uqcrh |
ubiquinol-cytochrome c reductase hinge protein |
994 |
0.31 |
| chr3_95740644_95740875 | 0.43 |
Ecm1 |
extracellular matrix protein 1 |
1190 |
0.25 |
| chr18_68228785_68229512 | 0.43 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
1081 |
0.51 |
| chr11_88840326_88840785 | 0.43 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
985 |
0.51 |
| chr6_118759013_118759164 | 0.43 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1114 |
0.62 |
| chr15_11741805_11742688 | 0.42 |
Gm41271 |
predicted gene, 41271 |
38543 |
0.18 |
| chr15_5555660_5555811 | 0.42 |
5430437J10Rik |
RIKEN cDNA 5430437J10 gene |
24477 |
0.23 |
| chr5_116456914_116457389 | 0.42 |
Gm42853 |
predicted gene 42853 |
333 |
0.83 |
| chr14_99366007_99366683 | 0.41 |
Gm49218 |
predicted gene, 49218 |
328 |
0.85 |
| chr13_83522624_83522907 | 0.41 |
Mef2c |
myocyte enhancer factor 2C |
708 |
0.79 |
| chr17_47909108_47910390 | 0.41 |
Gm15556 |
predicted gene 15556 |
12629 |
0.13 |
| chr3_53730067_53730769 | 0.41 |
Gm30735 |
predicted gene, 30735 |
4941 |
0.16 |
| chr3_63899898_63900049 | 0.41 |
Plch1 |
phospholipase C, eta 1 |
501 |
0.73 |
| chr4_134514105_134514256 | 0.40 |
Aunip |
aurora kinase A and ninein interacting protein |
3181 |
0.14 |
| chrX_138915472_138915880 | 0.40 |
Nrk |
Nik related kinase |
1245 |
0.61 |
| chr1_131232374_131233673 | 0.40 |
Rassf5 |
Ras association (RalGDS/AF-6) domain family member 5 |
552 |
0.67 |
| chrX_114816407_114816558 | 0.40 |
Gm14897 |
predicted gene 14897 |
41405 |
0.14 |
| chr5_65900728_65900879 | 0.39 |
Gm43769 |
predicted gene 43769 |
10821 |
0.11 |
| chr3_122243418_122243576 | 0.38 |
Gclm |
glutamate-cysteine ligase, modifier subunit |
2060 |
0.17 |
| chr18_64492934_64493085 | 0.37 |
Fech |
ferrochelatase |
2758 |
0.22 |
| chr14_30530906_30531057 | 0.37 |
Dcp1a |
decapping mRNA 1A |
7274 |
0.15 |
| chr10_42634471_42634622 | 0.37 |
Ostm1 |
osteopetrosis associated transmembrane protein 1 |
44370 |
0.13 |
| chr9_103364095_103364246 | 0.37 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
727 |
0.6 |
| chr3_155052033_155052416 | 0.37 |
Tnni3k |
TNNI3 interacting kinase |
3126 |
0.26 |
| chr2_155894612_155894870 | 0.37 |
Uqcc1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
15863 |
0.12 |
| chr4_132513569_132513723 | 0.36 |
Sesn2 |
sestrin 2 |
3145 |
0.12 |
| chr4_123990305_123991528 | 0.36 |
Gm12902 |
predicted gene 12902 |
64682 |
0.08 |
| chr1_135840170_135841177 | 0.35 |
Tnnt2 |
troponin T2, cardiac |
26 |
0.97 |
| chr5_106463547_106465134 | 0.35 |
Barhl2 |
BarH like homeobox 2 |
6174 |
0.18 |
| chr11_61142924_61143667 | 0.35 |
Tnfrsf13b |
tumor necrosis factor receptor superfamily, member 13b |
630 |
0.65 |
| chr19_6919975_6920251 | 0.34 |
Esrra |
estrogen related receptor, alpha |
938 |
0.29 |
| chr2_132427542_132428080 | 0.34 |
4921508D12Rik |
RIKEN cDNA 4921508D12 gene |
3070 |
0.25 |
| chr15_89168947_89170050 | 0.34 |
Plxnb2 |
plexin B2 |
1190 |
0.27 |
| chr1_135715343_135715494 | 0.33 |
Csrp1 |
cysteine and glycine-rich protein 1 |
4643 |
0.19 |
| chr6_14754259_14754642 | 0.33 |
Ppp1r3a |
protein phosphatase 1, regulatory subunit 3A |
824 |
0.77 |
| chrX_164437518_164437860 | 0.33 |
Asb11 |
ankyrin repeat and SOCS box-containing 11 |
369 |
0.85 |
| chr13_37859653_37859927 | 0.32 |
Rreb1 |
ras responsive element binding protein 1 |
1856 |
0.37 |
| chr8_27263249_27263470 | 0.32 |
Eif4ebp1 |
eukaryotic translation initiation factor 4E binding protein 1 |
1055 |
0.37 |
| chr8_39011566_39011717 | 0.32 |
Tusc3 |
tumor suppressor candidate 3 |
5731 |
0.26 |
| chr5_76778953_76779779 | 0.32 |
C530008M17Rik |
RIKEN cDNA C530008M17 gene |
26998 |
0.16 |
| chr3_35332632_35333522 | 0.32 |
Gm25442 |
predicted gene, 25442 |
7463 |
0.26 |
| chr16_96202794_96202945 | 0.32 |
Sh3bgr |
SH3-binding domain glutamic acid-rich protein |
2140 |
0.24 |
| chr14_20918784_20918935 | 0.32 |
Vcl |
vinculin |
10539 |
0.2 |
| chr18_54719777_54720132 | 0.32 |
Gm5821 |
predicted gene 5821 |
46178 |
0.16 |
| chr4_44663567_44664953 | 0.32 |
Gm12462 |
predicted gene 12462 |
155 |
0.94 |
| chr10_95297621_95297998 | 0.31 |
Gm48880 |
predicted gene, 48880 |
17044 |
0.14 |
| chr1_184675496_184676152 | 0.31 |
Gm38358 |
predicted gene, 38358 |
19210 |
0.14 |
| chr1_56974277_56974569 | 0.31 |
Satb2 |
special AT-rich sequence binding protein 2 |
1921 |
0.24 |
| chr17_30187860_30188415 | 0.31 |
Zfand3 |
zinc finger, AN1-type domain 3 |
9439 |
0.17 |
| chr9_64918332_64919221 | 0.30 |
Dennd4a |
DENN/MADD domain containing 4A |
6293 |
0.16 |
| chr2_38784133_38784675 | 0.30 |
Nr6a1 |
nuclear receptor subfamily 6, group A, member 1 |
65 |
0.95 |
| chr12_91383730_91384393 | 0.30 |
Cep128 |
centrosomal protein 128 |
292 |
0.68 |
| chr16_4519120_4519560 | 0.30 |
Srl |
sarcalumenin |
3723 |
0.19 |
| chr11_64656679_64656830 | 0.30 |
Gm24275 |
predicted gene, 24275 |
66114 |
0.13 |
| chr5_121184834_121185467 | 0.30 |
Gm42930 |
predicted gene 42930 |
154 |
0.93 |
| chr8_116919844_116920132 | 0.30 |
Cmc2 |
COX assembly mitochondrial protein 2 |
1377 |
0.33 |
| chrX_103472886_103473037 | 0.30 |
Xist |
inactive X specific transcripts |
5705 |
0.09 |
| chr14_69279140_69279312 | 0.30 |
Gm20236 |
predicted gene, 20236 |
2914 |
0.13 |
| chr12_69195806_69195957 | 0.30 |
Gm49383 |
predicted gene, 49383 |
1223 |
0.17 |
| chr11_99243687_99244404 | 0.29 |
Krt222 |
keratin 222 |
22 |
0.97 |
| chr12_100242288_100242784 | 0.29 |
Gm40557 |
predicted gene, 40557 |
92 |
0.96 |
| chr6_87422198_87422649 | 0.29 |
Bmp10 |
bone morphogenetic protein 10 |
6571 |
0.15 |
| chr13_30338441_30338963 | 0.29 |
Agtr1a |
angiotensin II receptor, type 1a |
2258 |
0.33 |
| chr8_46216441_46216738 | 0.29 |
Slc25a4 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
5305 |
0.12 |
| chr12_49387532_49388566 | 0.29 |
3110039M20Rik |
RIKEN cDNA 3110039M20 gene |
1603 |
0.26 |
| chr11_62629548_62630855 | 0.29 |
Lrrc75aos1 |
leucine rich repeat containing 75A, opposite strand 1 |
4503 |
0.09 |
| chr8_127552716_127553166 | 0.29 |
Gm6921 |
predicted pseudogene 6921 |
8494 |
0.23 |
| chr15_101191724_101192125 | 0.29 |
Mir6962 |
microRNA 6962 |
1945 |
0.2 |
| chr4_107004970_107005270 | 0.29 |
Gm12786 |
predicted gene 12786 |
17379 |
0.15 |
| chr18_69608508_69609200 | 0.29 |
Tcf4 |
transcription factor 4 |
3590 |
0.33 |
| chr3_58419356_58419775 | 0.28 |
Tsc22d2 |
TSC22 domain family, member 2 |
2081 |
0.31 |
| chr19_28580015_28580320 | 0.28 |
4933413C19Rik |
RIKEN cDNA 4933413C19 gene |
3127 |
0.3 |
| chr2_63982565_63982778 | 0.28 |
Fign |
fidgetin |
115317 |
0.07 |
| chr12_117743931_117744082 | 0.28 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
3411 |
0.26 |
| chr2_93008102_93008253 | 0.28 |
Prdm11 |
PR domain containing 11 |
5733 |
0.21 |
| chr15_77202946_77203567 | 0.28 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
180 |
0.94 |
| chr4_109677685_109677836 | 0.28 |
Faf1 |
Fas-associated factor 1 |
763 |
0.61 |
| chr5_106705019_106705272 | 0.28 |
Zfp644 |
zinc finger protein 644 |
7858 |
0.14 |
| chr1_155526845_155527426 | 0.27 |
Gm5532 |
predicted gene 5532 |
8 |
0.98 |
| chr15_40661942_40662224 | 0.27 |
Zfpm2 |
zinc finger protein, multitype 2 |
2907 |
0.38 |
| chr1_139459017_139459540 | 0.27 |
Aspm |
abnormal spindle microtubule assembly |
4459 |
0.2 |
| chrX_10992282_10993226 | 0.27 |
Gm14485 |
predicted gene 14485 |
28253 |
0.23 |
| chr7_109191327_109192334 | 0.27 |
Lmo1 |
LIM domain only 1 |
16623 |
0.17 |
| chr10_37988715_37989307 | 0.27 |
Gm24710 |
predicted gene, 24710 |
57331 |
0.15 |
| chr5_106757360_106758250 | 0.26 |
Gm8365 |
predicted gene 8365 |
19041 |
0.17 |
| chr1_190928081_190928453 | 0.26 |
Angel2 |
angel homolog 2 |
225 |
0.91 |
| chr15_40115274_40115425 | 0.26 |
9330182O14Rik |
RIKEN cDNA 9330182O14 gene |
19016 |
0.17 |
| chr4_151083338_151083489 | 0.26 |
Camta1 |
calmodulin binding transcription activator 1 |
3620 |
0.2 |
| chr8_106890881_106891378 | 0.26 |
Chtf8 |
CTF8, chromosome transmission fidelity factor 8 |
2086 |
0.18 |
| chr5_99970736_99971131 | 0.25 |
Hnrnpd |
heterogeneous nuclear ribonucleoprotein D |
4546 |
0.16 |
| chr2_133739629_133739780 | 0.25 |
Gm25258 |
predicted gene, 25258 |
155283 |
0.04 |
| chr6_86683779_86684487 | 0.25 |
Snrnp27 |
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
358 |
0.77 |
| chrY_3871382_3871533 | 0.25 |
Gm18177 |
predicted gene, 18177 |
4471 |
0.13 |
| chr5_75083070_75083610 | 0.25 |
Gsx2 |
GS homeobox 2 |
7738 |
0.12 |
| chr15_85463399_85463550 | 0.25 |
7530416G11Rik |
RIKEN cDNA 7530416G11 gene |
39753 |
0.14 |
| chr15_9529382_9529634 | 0.25 |
Il7r |
interleukin 7 receptor |
258 |
0.94 |
| chr18_42252337_42252647 | 0.25 |
Rps19-ps4 |
ribosomal protein S19, pseudogene 4 |
94 |
0.95 |
| chr12_101978605_101978756 | 0.25 |
Ndufb1-ps |
NADH:ubiquinone oxidoreductase subunit B1 |
1612 |
0.3 |
| chr3_135421547_135421698 | 0.25 |
Cisd2 |
CDGSH iron sulfur domain 2 |
1457 |
0.26 |
| chr17_40811481_40812037 | 0.25 |
Rhag |
Rhesus blood group-associated A glycoprotein |
575 |
0.7 |
| chr4_131844041_131844267 | 0.24 |
Mecr |
mitochondrial trans-2-enoyl-CoA reductase |
649 |
0.61 |
| chr17_10293681_10293832 | 0.24 |
Qk |
quaking |
10823 |
0.25 |
| chr17_14959778_14960235 | 0.24 |
Phf10 |
PHD finger protein 10 |
479 |
0.69 |
| chr11_117414820_117416356 | 0.24 |
Gm11729 |
predicted gene 11729 |
1199 |
0.39 |
| chr12_56298063_56298592 | 0.24 |
Gm47682 |
predicted gene, 47682 |
43929 |
0.1 |
| chr2_119619277_119620230 | 0.24 |
Nusap1 |
nucleolar and spindle associated protein 1 |
1024 |
0.3 |
| chr16_23780715_23780866 | 0.24 |
Gm49515 |
predicted gene, 49515 |
663 |
0.75 |
| chr18_56715299_56715450 | 0.24 |
Lmnb1 |
lamin B1 |
7561 |
0.19 |
| chr19_34165708_34165981 | 0.24 |
Ankrd22 |
ankyrin repeat domain 22 |
209 |
0.93 |
| chr8_81856300_81856587 | 0.23 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
75 |
0.98 |
| chr9_77344708_77344864 | 0.23 |
Mlip |
muscular LMNA-interacting protein |
3007 |
0.22 |
| chr7_69891593_69891960 | 0.23 |
Gm44690 |
predicted gene 44690 |
7416 |
0.25 |
| chr3_9573681_9573832 | 0.23 |
Zfp704 |
zinc finger protein 704 |
8457 |
0.25 |
| chr8_33916300_33917561 | 0.23 |
Rbpms |
RNA binding protein gene with multiple splicing |
12346 |
0.17 |
| chr16_92478627_92479097 | 0.23 |
2410124H12Rik |
RIKEN cDNA 2410124H12 gene |
120 |
0.95 |
| chr13_76999543_76999694 | 0.23 |
Slf1 |
SMC5-SMC6 complex localization factor 1 |
47779 |
0.16 |
| chr4_40851422_40852119 | 0.23 |
Gm25931 |
predicted gene, 25931 |
1368 |
0.21 |
| chrX_143825863_143827628 | 0.23 |
Capn6 |
calpain 6 |
587 |
0.46 |
| chr14_47299300_47299451 | 0.23 |
Mapk1ip1l |
mitogen-activated protein kinase 1 interacting protein 1-like |
604 |
0.41 |
| chr8_48596222_48596652 | 0.23 |
Gm45772 |
predicted gene 45772 |
34732 |
0.19 |
| chr12_9253584_9253735 | 0.23 |
Gm46323 |
predicted gene, 46323 |
117779 |
0.06 |
| chr1_127676820_127678197 | 0.23 |
Tmem163 |
transmembrane protein 163 |
513 |
0.83 |
| chr7_113249773_113249924 | 0.23 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
10094 |
0.2 |
| chr12_84589686_84590923 | 0.23 |
Abcd4 |
ATP-binding cassette, sub-family D (ALD), member 4 |
13748 |
0.15 |
| chr2_58509453_58510324 | 0.23 |
Acvr1 |
activin A receptor, type 1 |
6384 |
0.23 |
| chr16_33060324_33060635 | 0.22 |
Lmln |
leishmanolysin-like (metallopeptidase M8 family) |
2042 |
0.19 |
| chr8_119490510_119491264 | 0.22 |
Slc38a8 |
solute carrier family 38, member 8 |
4008 |
0.17 |
| chr7_6281481_6281771 | 0.22 |
Gm45069 |
predicted gene 45069 |
1984 |
0.19 |
| chr17_44136324_44136532 | 0.22 |
Clic5 |
chloride intracellular channel 5 |
1660 |
0.43 |
| chr6_24599678_24600100 | 0.22 |
Lmod2 |
leiomodin 2 (cardiac) |
2127 |
0.25 |
| chr13_12033129_12034063 | 0.22 |
Gm47493 |
predicted gene, 47493 |
18459 |
0.21 |
| chr6_145276027_145276999 | 0.22 |
Rps25-ps1 |
ribosomal protein S25, pseudogene 1 |
10113 |
0.1 |
| chrX_16816049_16816282 | 0.22 |
Maob |
monoamine oxidase B |
1110 |
0.65 |
| chr10_42613700_42614446 | 0.22 |
Ostm1 |
osteopetrosis associated transmembrane protein 1 |
30235 |
0.16 |
| chr3_21935032_21935297 | 0.22 |
Gm43674 |
predicted gene 43674 |
63304 |
0.12 |
| chr13_94595583_94596884 | 0.22 |
Gm36991 |
predicted gene, 36991 |
23047 |
0.16 |
| chr2_31692679_31692830 | 0.22 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
3143 |
0.14 |
| chr3_70104094_70104909 | 0.22 |
Gm37585 |
predicted gene, 37585 |
12031 |
0.22 |
| chr3_66982218_66983084 | 0.22 |
Shox2 |
short stature homeobox 2 |
880 |
0.47 |
| chr7_75781306_75781612 | 0.21 |
AU020206 |
expressed sequence AU020206 |
640 |
0.66 |
| chr12_50111378_50111869 | 0.21 |
Gm40418 |
predicted gene, 40418 |
8686 |
0.33 |
| chr2_20966839_20967346 | 0.21 |
Arhgap21 |
Rho GTPase activating protein 21 |
628 |
0.68 |
| chr10_61413178_61413819 | 0.21 |
Nodal |
nodal |
4474 |
0.13 |
| chr6_45096373_45096524 | 0.21 |
Gm43879 |
predicted gene, 43879 |
15702 |
0.16 |
| chr10_31386394_31386545 | 0.21 |
Gm48094 |
predicted gene, 48094 |
12454 |
0.13 |
| chr9_72791334_72791686 | 0.21 |
Gm27204 |
predicted gene 27204 |
15285 |
0.11 |
| chr6_5392568_5392891 | 0.21 |
Asb4 |
ankyrin repeat and SOCS box-containing 4 |
2302 |
0.38 |
| chr10_122388056_122388209 | 0.21 |
Gm36041 |
predicted gene, 36041 |
1240 |
0.53 |
| chr10_121625316_121626404 | 0.21 |
Xpot |
exportin, tRNA (nuclear export receptor for tRNAs) |
456 |
0.76 |
| chr12_69157543_69157739 | 0.21 |
Rps29 |
ribosomal protein S29 |
1413 |
0.15 |
| chr9_25150579_25151803 | 0.21 |
Herpud2 |
HERPUD family member 2 |
507 |
0.62 |
| chr3_53465387_53466092 | 0.21 |
Proser1 |
proline and serine rich 1 |
1807 |
0.22 |
| chr14_54362334_54362485 | 0.21 |
Oxa1l |
oxidase assembly 1-like |
1364 |
0.25 |
| chr9_87406338_87406888 | 0.21 |
Gm9540 |
predicted gene 9540 |
12363 |
0.26 |
| chr9_67261068_67261449 | 0.21 |
Tln2 |
talin 2 |
835 |
0.67 |
| chr15_80784467_80785284 | 0.21 |
Tnrc6b |
trinucleotide repeat containing 6b |
13767 |
0.2 |
| chr17_23541898_23542685 | 0.21 |
6330415G19Rik |
RIKEN cDNA 6330415G19 gene |
8508 |
0.09 |
| chr15_40656769_40657228 | 0.21 |
Zfpm2 |
zinc finger protein, multitype 2 |
1647 |
0.51 |
| chr3_69008816_69009141 | 0.20 |
Mir15b |
microRNA 15b |
794 |
0.37 |
| chr6_18849259_18849471 | 0.20 |
Lsm8 |
LSM8 homolog, U6 small nuclear RNA associated |
697 |
0.51 |
| chr7_109115172_109115418 | 0.20 |
Gm44781 |
predicted gene 44781 |
27592 |
0.13 |
| chr15_25773704_25775483 | 0.20 |
Myo10 |
myosin X |
606 |
0.78 |
| chr4_145585026_145585604 | 0.20 |
Gm13212 |
predicted gene 13212 |
112 |
0.93 |
| chr16_30236013_30236491 | 0.20 |
Gm49645 |
predicted gene, 49645 |
18900 |
0.13 |
| chr3_59028229_59029161 | 0.20 |
Med12l |
mediator complex subunit 12-like |
21717 |
0.13 |
| chr5_103424052_103424207 | 0.20 |
Ptpn13 |
protein tyrosine phosphatase, non-receptor type 13 |
1063 |
0.56 |
| chr14_45330987_45331140 | 0.20 |
Psmc6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
308 |
0.85 |
| chr18_58206037_58206198 | 0.20 |
Fbn2 |
fibrillin 2 |
3809 |
0.3 |
| chr2_6719607_6719924 | 0.20 |
Celf2 |
CUGBP, Elav-like family member 2 |
1848 |
0.48 |
| chr10_92439233_92439384 | 0.20 |
4930401A07Rik |
RIKEN cDNA 4930401A07 gene |
38294 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.4 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.5 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 | 0.4 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.3 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0035912 | dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.0 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.0 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.0 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0060379 | cardiac muscle cell myoblast differentiation(GO:0060379) |
| 0.0 | 0.0 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0070694 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |