Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
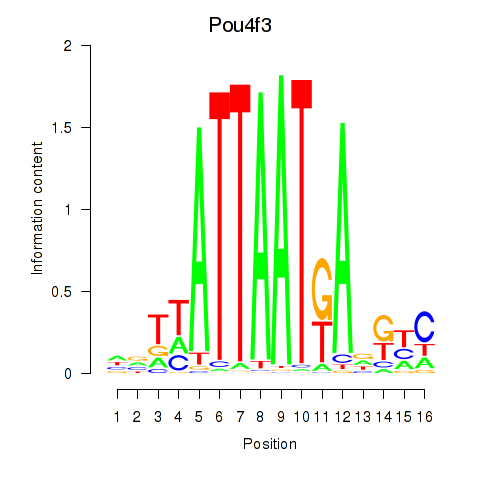
Results for Pou4f3
Z-value: 0.80
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSMUSG00000024497.3 | POU domain, class 4, transcription factor 3 |
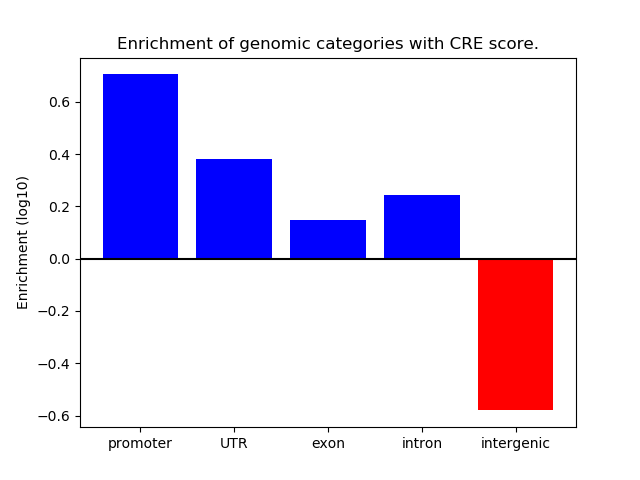
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_42394146_42396205 | Pou4f3 | 100 | 0.969657 | 0.40 | 1.4e-03 | Click! |
| chr18_42398744_42398916 | Pou4f3 | 3755 | 0.231122 | 0.32 | 1.3e-02 | Click! |
| chr18_42400574_42400820 | Pou4f3 | 5622 | 0.205079 | 0.27 | 3.6e-02 | Click! |
| chr18_42396624_42396775 | Pou4f3 | 1624 | 0.381433 | 0.27 | 3.8e-02 | Click! |
| chr18_42399066_42399299 | Pou4f3 | 4107 | 0.223771 | 0.26 | 4.9e-02 | Click! |
Activity of the Pou4f3 motif across conditions
Conditions sorted by the z-value of the Pou4f3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
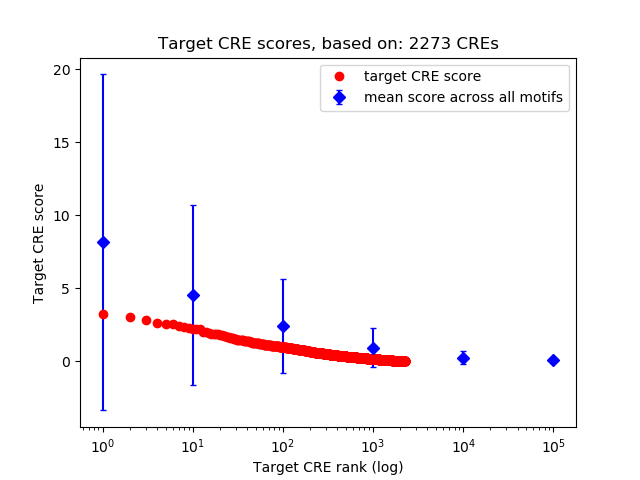
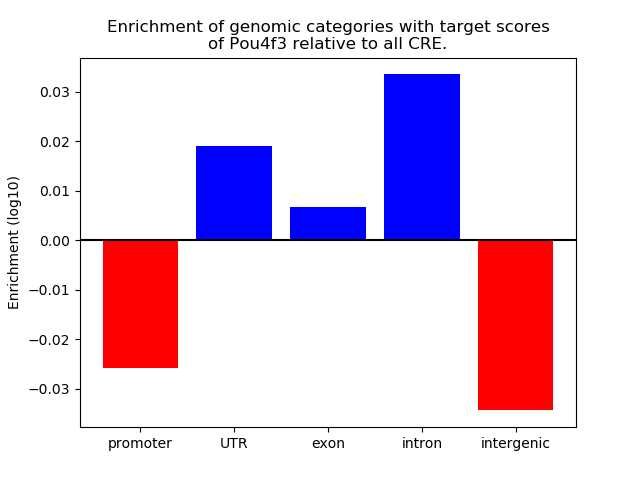
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_29143400_29144848 | 3.23 |
Soga3 |
SOGA family member 3 |
65 |
0.5 |
| chr8_54956010_54956394 | 3.03 |
Gpm6a |
glycoprotein m6a |
1359 |
0.38 |
| chr4_110290101_110291006 | 2.82 |
Elavl4 |
ELAV like RNA binding protein 4 |
281 |
0.95 |
| chr4_33926104_33927188 | 2.61 |
Cnr1 |
cannabinoid receptor 1 (brain) |
444 |
0.88 |
| chr13_83736071_83736534 | 2.56 |
Gm33366 |
predicted gene, 33366 |
2233 |
0.18 |
| chr14_64591760_64591911 | 2.53 |
Mir3078 |
microRNA 3078 |
650 |
0.54 |
| chr7_87586513_87587584 | 2.45 |
Grm5 |
glutamate receptor, metabotropic 5 |
2650 |
0.4 |
| chr18_59062200_59063436 | 2.34 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
307 |
0.94 |
| chr2_65620767_65621991 | 2.30 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
568 |
0.82 |
| chr8_123412815_123413352 | 2.25 |
Tubb3 |
tubulin, beta 3 class III |
1493 |
0.18 |
| chr6_55678280_55679200 | 2.24 |
Neurod6 |
neurogenic differentiation 6 |
2523 |
0.32 |
| chr2_178144058_178144423 | 2.21 |
Phactr3 |
phosphatase and actin regulator 3 |
2307 |
0.37 |
| chr8_40633582_40633735 | 2.04 |
Mtmr7 |
myotubularin related protein 7 |
1101 |
0.48 |
| chrX_134406275_134406692 | 2.02 |
Drp2 |
dystrophin related protein 2 |
1681 |
0.35 |
| chr1_172056022_172057415 | 1.97 |
Nhlh1 |
nescient helix loop helix 1 |
855 |
0.45 |
| chr12_29537463_29537646 | 1.88 |
Myt1l |
myelin transcription factor 1-like |
2332 |
0.32 |
| chr13_70840476_70842094 | 1.86 |
Adamts16 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 16 |
490 |
0.63 |
| chr1_81077232_81078427 | 1.85 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
246 |
0.96 |
| chr10_90578974_90579573 | 1.84 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
2281 |
0.42 |
| chr14_55056074_55056891 | 1.77 |
Gm20687 |
predicted gene 20687 |
989 |
0.3 |
| chr2_77701567_77703605 | 1.77 |
Zfp385b |
zinc finger protein 385B |
686 |
0.8 |
| chr12_88723574_88724074 | 1.74 |
Nrxn3 |
neurexin III |
715 |
0.73 |
| chr4_33927284_33927435 | 1.70 |
Cnr1 |
cannabinoid receptor 1 (brain) |
1157 |
0.6 |
| chr8_108535196_108536010 | 1.67 |
Gm39244 |
predicted gene, 39244 |
1344 |
0.54 |
| chr3_86543379_86544222 | 1.67 |
Lrba |
LPS-responsive beige-like anchor |
1767 |
0.38 |
| chr2_14875285_14875631 | 1.62 |
Cacnb2 |
calcium channel, voltage-dependent, beta 2 subunit |
706 |
0.69 |
| chr11_119544322_119545100 | 1.59 |
Nptx1 |
neuronal pentraxin 1 |
3042 |
0.19 |
| chr5_37241461_37244349 | 1.57 |
Crmp1 |
collapsin response mediator protein 1 |
171 |
0.95 |
| chr2_82056608_82056868 | 1.52 |
Zfp804a |
zinc finger protein 804A |
3516 |
0.36 |
| chr11_96338771_96339811 | 1.50 |
Hoxb3 |
homeobox B3 |
1654 |
0.16 |
| chr1_137901653_137901959 | 1.49 |
Gm4258 |
predicted gene 4258 |
3208 |
0.12 |
| chr9_91357788_91358422 | 1.47 |
Zic4 |
zinc finger protein of the cerebellum 4 |
4308 |
0.12 |
| chr2_51599699_51600378 | 1.47 |
Gm13491 |
predicted gene 13491 |
4310 |
0.24 |
| chr2_65929929_65930575 | 1.47 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
115 |
0.97 |
| chrX_153501207_153502250 | 1.45 |
Ubqln2 |
ubiquilin 2 |
3501 |
0.22 |
| chr7_49907312_49908741 | 1.43 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
2120 |
0.4 |
| chrX_160992631_160993204 | 1.41 |
Cdkl5 |
cyclin-dependent kinase-like 5 |
1787 |
0.46 |
| chr12_71048832_71049275 | 1.40 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
712 |
0.65 |
| chr8_48508743_48509680 | 1.39 |
Tenm3 |
teneurin transmembrane protein 3 |
46102 |
0.19 |
| chrX_110811626_110812334 | 1.37 |
Gm44593 |
predicted gene 44593 |
344 |
0.89 |
| chr12_29529828_29531185 | 1.37 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr10_90832070_90832464 | 1.36 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
2424 |
0.26 |
| chr8_41052368_41053980 | 1.34 |
Gm16193 |
predicted gene 16193 |
64 |
0.96 |
| chr13_44842150_44842855 | 1.34 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
1719 |
0.39 |
| chr10_87490860_87491182 | 1.32 |
Ascl1 |
achaete-scute family bHLH transcription factor 1 |
2639 |
0.28 |
| chr10_12611642_12612156 | 1.28 |
Utrn |
utrophin |
2932 |
0.38 |
| chr8_34890130_34891317 | 1.26 |
Tnks |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
572 |
0.8 |
| chr14_66865047_66865736 | 1.24 |
Dpysl2 |
dihydropyrimidinase-like 2 |
3297 |
0.19 |
| chr3_38894285_38895428 | 1.23 |
Fat4 |
FAT atypical cadherin 4 |
3914 |
0.27 |
| chr12_107990188_107992301 | 1.23 |
Bcl11b |
B cell leukemia/lymphoma 11B |
12170 |
0.28 |
| chr9_41584760_41585051 | 1.23 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
2864 |
0.14 |
| chr12_76081146_76082447 | 1.23 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
140 |
0.97 |
| chr1_42703141_42704653 | 1.22 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
4155 |
0.16 |
| chr10_96621972_96622429 | 1.22 |
Btg1 |
BTG anti-proliferation factor 1 |
4646 |
0.23 |
| chr1_9296437_9296701 | 1.22 |
Sntg1 |
syntrophin, gamma 1 |
1667 |
0.35 |
| chr18_69500231_69501482 | 1.21 |
Tcf4 |
transcription factor 4 |
20 |
0.99 |
| chr19_14593738_14594104 | 1.20 |
Tle4 |
transducin-like enhancer of split 4 |
1618 |
0.51 |
| chr14_32603268_32603785 | 1.19 |
Prrxl1 |
paired related homeobox protein-like 1 |
3568 |
0.21 |
| chr6_8948312_8949000 | 1.19 |
Nxph1 |
neurexophilin 1 |
42 |
0.99 |
| chr17_83792570_83792796 | 1.19 |
Mta3 |
metastasis associated 3 |
2739 |
0.29 |
| chr13_115346333_115347310 | 1.16 |
Gm47891 |
predicted gene, 47891 |
64620 |
0.13 |
| chr12_72233267_72233815 | 1.15 |
Rtn1 |
reticulon 1 |
2198 |
0.35 |
| chr5_70842167_70842810 | 1.14 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
129 |
0.98 |
| chr15_85682337_85682535 | 1.14 |
Lncppara |
long noncoding RNA near Ppara |
21337 |
0.12 |
| chr4_110281271_110281426 | 1.13 |
Elavl4 |
ELAV like RNA binding protein 4 |
5268 |
0.33 |
| chr14_55052408_55052978 | 1.12 |
Zfhx2os |
zinc finger homeobox 2, opposite strand |
1176 |
0.24 |
| chr11_32001099_32002296 | 1.11 |
Nsg2 |
neuron specific gene family member 2 |
1195 |
0.52 |
| chr15_44746196_44747245 | 1.10 |
Sybu |
syntabulin (syntaxin-interacting) |
1068 |
0.56 |
| chr17_15371644_15373099 | 1.10 |
Dll1 |
delta like canonical Notch ligand 1 |
540 |
0.73 |
| chr2_92399434_92399703 | 1.10 |
Mapk8ip1 |
mitogen-activated protein kinase 8 interacting protein 1 |
1695 |
0.21 |
| chr4_31880335_31880706 | 1.09 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
83577 |
0.09 |
| chr5_98183897_98184048 | 1.09 |
A730035I17Rik |
RIKEN cDNA A730035I17 gene |
1238 |
0.41 |
| chr2_136062179_136062393 | 1.08 |
Lamp5 |
lysosomal-associated membrane protein family, member 5 |
2855 |
0.3 |
| chr13_83735228_83735549 | 1.08 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2822 |
0.16 |
| chr2_123533004_123533690 | 1.08 |
Gm13988 |
predicted gene 13988 |
259423 |
0.02 |
| chr10_70484934_70485670 | 1.06 |
Gm29783 |
predicted gene, 29783 |
19946 |
0.19 |
| chr15_4375207_4376518 | 1.05 |
Plcxd3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
358 |
0.93 |
| chr12_3368013_3368276 | 1.05 |
Gm48511 |
predicted gene, 48511 |
285 |
0.85 |
| chr3_86544323_86545126 | 1.05 |
Lrba |
LPS-responsive beige-like anchor |
2031 |
0.33 |
| chr1_89308745_89309143 | 1.05 |
Gm28342 |
predicted gene 28342 |
49200 |
0.14 |
| chr17_32162253_32163139 | 1.04 |
Notch3 |
notch 3 |
4156 |
0.15 |
| chr12_33225182_33225605 | 1.04 |
Atxn7l1os1 |
ataxin 7-like 1, opposite strand 1 |
3461 |
0.27 |
| chr1_32173438_32173903 | 1.04 |
Khdrbs2 |
KH domain containing, RNA binding, signal transduction associated 2 |
783 |
0.74 |
| chr11_96328165_96328613 | 1.04 |
Hoxb3 |
homeobox B3 |
327 |
0.72 |
| chr2_157916786_157917265 | 1.03 |
Vstm2l |
V-set and transmembrane domain containing 2-like |
2372 |
0.3 |
| chr17_62746805_62747199 | 1.03 |
Efna5 |
ephrin A5 |
134142 |
0.05 |
| chr8_125897573_125897839 | 1.03 |
A730098A19Rik |
RIKEN cDNA A730098A19 gene |
208 |
0.8 |
| chr2_3168763_3169231 | 1.03 |
Fam171a1 |
family with sequence similarity 171, member A1 |
50312 |
0.13 |
| chr9_100266506_100267433 | 1.02 |
Gm28167 |
predicted gene 28167 |
56555 |
0.13 |
| chr3_134236641_134237783 | 1.02 |
Cxxc4 |
CXXC finger 4 |
392 |
0.78 |
| chr4_82494042_82494837 | 1.01 |
Nfib |
nuclear factor I/B |
4877 |
0.23 |
| chr7_25004827_25006284 | 1.01 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
340 |
0.79 |
| chr18_35212708_35213458 | 1.01 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
1847 |
0.28 |
| chr18_69352477_69352948 | 1.01 |
Tcf4 |
transcription factor 4 |
3768 |
0.32 |
| chr2_153440322_153442352 | 1.00 |
Nol4l |
nucleolar protein 4-like |
3142 |
0.22 |
| chr13_8207058_8207829 | 1.00 |
Adarb2 |
adenosine deaminase, RNA-specific, B2 |
4521 |
0.2 |
| chr15_103058659_103059955 | 1.00 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
1 |
0.95 |
| chr15_99056560_99057587 | 0.99 |
Prph |
peripherin |
1103 |
0.3 |
| chr6_15196934_15197697 | 0.99 |
Foxp2 |
forkhead box P2 |
351 |
0.94 |
| chr16_7041535_7042315 | 0.99 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
27921 |
0.28 |
| chr18_31316134_31317386 | 0.99 |
Rit2 |
Ras-like without CAAX 2 |
351 |
0.9 |
| chr1_169745785_169746010 | 0.98 |
Rgs4 |
regulator of G-protein signaling 4 |
1726 |
0.41 |
| chr13_44100281_44100444 | 0.97 |
Gm33630 |
predicted gene, 33630 |
402 |
0.85 |
| chr15_78115657_78115889 | 0.97 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
3933 |
0.18 |
| chr9_74869680_74869831 | 0.96 |
Onecut1 |
one cut domain, family member 1 |
3271 |
0.2 |
| chr3_115774354_115774995 | 0.96 |
Gm9889 |
predicted gene 9889 |
59524 |
0.1 |
| chr3_35481945_35482453 | 0.95 |
Gm7733 |
predicted gene 7733 |
55601 |
0.12 |
| chr1_66324716_66324867 | 0.95 |
Map2 |
microtubule-associated protein 2 |
2689 |
0.25 |
| chr6_13832197_13832750 | 0.94 |
Gpr85 |
G protein-coupled receptor 85 |
4768 |
0.23 |
| chr11_111605019_111605670 | 0.94 |
Gm11676 |
predicted gene 11676 |
7962 |
0.32 |
| chr2_151631540_151632560 | 0.94 |
Snph |
syntaphilin |
421 |
0.78 |
| chr8_106985656_106985807 | 0.93 |
Gm22085 |
predicted gene, 22085 |
23880 |
0.1 |
| chr13_15486652_15487073 | 0.93 |
Gli3 |
GLI-Kruppel family member GLI3 |
22882 |
0.17 |
| chr13_99031199_99031610 | 0.93 |
Gm47057 |
predicted gene, 47057 |
3444 |
0.18 |
| chr16_77236731_77239778 | 0.92 |
Mir99ahg |
Mir99a and Mirlet7c-1 host gene (non-protein coding) |
1935 |
0.4 |
| chr8_114911140_114911535 | 0.91 |
Gm22556 |
predicted gene, 22556 |
141576 |
0.05 |
| chr12_99340176_99340397 | 0.91 |
3300002A11Rik |
RIKEN cDNA 3300002A11 gene |
868 |
0.5 |
| chr18_72149524_72149675 | 0.90 |
Dcc |
deleted in colorectal carcinoma |
201418 |
0.03 |
| chr10_91581035_91581404 | 0.90 |
Gm47091 |
predicted gene, 47091 |
59671 |
0.14 |
| chr2_65668738_65669312 | 0.90 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
260 |
0.95 |
| chr3_55786102_55786728 | 0.90 |
Nbea |
neurobeachin |
487 |
0.8 |
| chr7_35848412_35848964 | 0.89 |
Gm28514 |
predicted gene 28514 |
10428 |
0.21 |
| chr11_77485097_77486316 | 0.89 |
Ankrd13b |
ankyrin repeat domain 13b |
3960 |
0.14 |
| chr8_90816068_90816342 | 0.89 |
Gm19935 |
predicted gene, 19935 |
1188 |
0.38 |
| chr2_165367693_165368982 | 0.88 |
Zfp663 |
zinc finger protein 663 |
386 |
0.8 |
| chr1_6732598_6732857 | 0.88 |
St18 |
suppression of tumorigenicity 18 |
2143 |
0.43 |
| chr13_97243359_97243510 | 0.88 |
Enc1 |
ectodermal-neural cortex 1 |
2329 |
0.26 |
| chr18_36930202_36930714 | 0.87 |
Pcdha1 |
protocadherin alpha 1 |
173 |
0.84 |
| chr8_78433398_78433999 | 0.87 |
Pou4f2 |
POU domain, class 4, transcription factor 2 |
2947 |
0.27 |
| chr3_144318844_144319150 | 0.87 |
Gm43447 |
predicted gene 43447 |
1217 |
0.51 |
| chr15_77150114_77150989 | 0.87 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3005 |
0.19 |
| chr10_45483988_45484272 | 0.86 |
Lin28b |
lin-28 homolog B (C. elegans) |
1647 |
0.42 |
| chr13_104111586_104112312 | 0.86 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
2120 |
0.26 |
| chr7_36703759_36704512 | 0.86 |
Tshz3 |
teashirt zinc finger family member 3 |
5918 |
0.14 |
| chr17_66444861_66445266 | 0.86 |
Mtcl1 |
microtubule crosslinking factor 1 |
3306 |
0.2 |
| chr14_31778025_31778176 | 0.85 |
Ankrd28 |
ankyrin repeat domain 28 |
2293 |
0.27 |
| chr1_118434945_118435685 | 0.85 |
Clasp1 |
CLIP associating protein 1 |
15591 |
0.13 |
| chr19_55897010_55897961 | 0.85 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
824 |
0.72 |
| chr8_47240952_47241121 | 0.84 |
Stox2 |
storkhead box 2 |
1301 |
0.49 |
| chr13_28881136_28881895 | 0.84 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
1941 |
0.32 |
| chr11_96309485_96310411 | 0.84 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
3038 |
0.09 |
| chr8_109310721_109311190 | 0.84 |
Gm1943 |
predicted gene 1943 |
29909 |
0.2 |
| chr9_34488424_34489634 | 0.84 |
Kirrel3 |
kirre like nephrin family adhesion molecule 3 |
93 |
0.98 |
| chr4_114842209_114842913 | 0.83 |
Gm23230 |
predicted gene, 23230 |
2556 |
0.25 |
| chr13_78937886_78938133 | 0.82 |
Gm8345 |
predicted gene 8345 |
16783 |
0.29 |
| chr13_69734884_69735178 | 0.82 |
Ube2ql1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
4858 |
0.14 |
| chr8_67817715_67818694 | 0.82 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
29 |
0.99 |
| chr15_98952050_98952507 | 0.81 |
Gm49450 |
predicted gene, 49450 |
1273 |
0.2 |
| chr11_64656679_64656830 | 0.81 |
Gm24275 |
predicted gene, 24275 |
66114 |
0.13 |
| chr17_66761376_66761829 | 0.80 |
Gm49939 |
predicted gene, 49939 |
17279 |
0.14 |
| chr15_78854800_78855024 | 0.80 |
Lgals2 |
lectin, galactose-binding, soluble 2 |
617 |
0.51 |
| chr1_136228373_136230942 | 0.79 |
Inava |
innate immunity activator |
362 |
0.76 |
| chr7_136196592_136196867 | 0.79 |
Gm36737 |
predicted gene, 36737 |
23433 |
0.22 |
| chr5_65133569_65133946 | 0.79 |
Klhl5 |
kelch-like 5 |
2086 |
0.25 |
| chr6_100270793_100270944 | 0.79 |
Gm44044 |
predicted gene, 44044 |
2830 |
0.23 |
| chr8_90958942_90959524 | 0.79 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
3798 |
0.21 |
| chr11_24090958_24091311 | 0.79 |
Bcl11a |
B cell CLL/lymphoma 11A (zinc finger protein) |
10464 |
0.14 |
| chr6_36797061_36797436 | 0.79 |
Ptn |
pleiotrophin |
12931 |
0.26 |
| chr16_31254473_31254624 | 0.79 |
Gm27415 |
predicted gene, 27415 |
6957 |
0.12 |
| chr3_141338481_141339046 | 0.78 |
Pdha2 |
pyruvate dehydrogenase E1 alpha 2 |
126408 |
0.05 |
| chr10_87500739_87501897 | 0.78 |
Gm48120 |
predicted gene, 48120 |
6544 |
0.19 |
| chr1_189932345_189932517 | 0.78 |
Smyd2 |
SET and MYND domain containing 2 |
10068 |
0.18 |
| chr1_172341079_172341970 | 0.77 |
Kcnj10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
314 |
0.81 |
| chr14_75964008_75964248 | 0.77 |
Gm25517 |
predicted gene, 25517 |
8491 |
0.18 |
| chr5_120409346_120410480 | 0.77 |
Lhx5 |
LIM homeobox protein 5 |
21786 |
0.12 |
| chr11_96336581_96337686 | 0.77 |
Hoxb3 |
homeobox B3 |
3812 |
0.09 |
| chr2_102448665_102449240 | 0.77 |
Fjx1 |
four jointed box 1 |
3547 |
0.29 |
| chr8_109250884_109251908 | 0.77 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
1530 |
0.52 |
| chr17_78205213_78205510 | 0.76 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
4296 |
0.17 |
| chr2_161510838_161511124 | 0.76 |
Gm29229 |
predicted gene 29229 |
3031 |
0.32 |
| chr7_36704897_36705048 | 0.76 |
Gm37452 |
predicted gene, 37452 |
5202 |
0.14 |
| chr12_67218771_67218992 | 0.75 |
Mdga2 |
MAM domain containing glycosylphosphatidylinositol anchor 2 |
2161 |
0.43 |
| chr5_9623201_9623446 | 0.74 |
Gm42455 |
predicted gene 42455 |
20716 |
0.24 |
| chr6_146892977_146893128 | 0.74 |
Ppfibp1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
4463 |
0.18 |
| chrX_69363262_69363944 | 0.74 |
Gm14705 |
predicted gene 14705 |
2334 |
0.33 |
| chr3_55464042_55464726 | 0.74 |
Dclk1 |
doublecortin-like kinase 1 |
2412 |
0.23 |
| chr8_88793832_88794104 | 0.73 |
Rps6-ps2 |
ribosomal protein S6, pseudogene 2 |
12423 |
0.2 |
| chr10_111925331_111925637 | 0.73 |
Gm47880 |
predicted gene, 47880 |
36470 |
0.11 |
| chr11_69834618_69835201 | 0.73 |
Nlgn2 |
neuroligin 2 |
60 |
0.9 |
| chr19_20009817_20010437 | 0.72 |
Gm22684 |
predicted gene, 22684 |
23508 |
0.22 |
| chr1_64116857_64117480 | 0.72 |
Klf7 |
Kruppel-like factor 7 (ubiquitous) |
4314 |
0.23 |
| chr2_31638722_31641540 | 0.72 |
Prdm12 |
PR domain containing 12 |
94 |
0.84 |
| chr10_94941542_94941964 | 0.72 |
Plxnc1 |
plexin C1 |
3082 |
0.28 |
| chr9_47533054_47533827 | 0.72 |
Cadm1 |
cell adhesion molecule 1 |
3067 |
0.25 |
| chr18_25548926_25549140 | 0.71 |
Celf4 |
CUGBP, Elav-like family member 4 |
47794 |
0.16 |
| chr1_143644977_143645827 | 0.71 |
Cdc73 |
cell division cycle 73, Paf1/RNA polymerase II complex component |
2877 |
0.24 |
| chr3_141467853_141468214 | 0.71 |
Unc5c |
unc-5 netrin receptor C |
2362 |
0.32 |
| chr10_99247088_99247239 | 0.71 |
Gm48884 |
predicted gene, 48884 |
9547 |
0.11 |
| chr17_91091821_91092017 | 0.70 |
Nrxn1 |
neurexin I |
814 |
0.55 |
| chr8_31915784_31916390 | 0.70 |
Nrg1 |
neuregulin 1 |
1563 |
0.42 |
| chr12_26533196_26533974 | 0.70 |
Gm46344 |
predicted gene, 46344 |
46794 |
0.11 |
| chr4_130568948_130569534 | 0.70 |
Nkain1 |
Na+/K+ transporting ATPase interacting 1 |
4725 |
0.27 |
| chr3_152106774_152107680 | 0.69 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
195 |
0.8 |
| chr16_67618825_67619284 | 0.69 |
Cadm2 |
cell adhesion molecule 2 |
1439 |
0.51 |
| chr1_177199086_177199237 | 0.69 |
Gm37706 |
predicted gene, 37706 |
28299 |
0.14 |
| chr15_98989928_98991865 | 0.69 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
5002 |
0.1 |
| chr1_42286601_42286752 | 0.68 |
Gm28175 |
predicted gene 28175 |
24819 |
0.18 |
| chr9_56159774_56159955 | 0.68 |
Tspan3 |
tetraspanin 3 |
678 |
0.68 |
| chr2_105675959_105678109 | 0.67 |
Pax6 |
paired box 6 |
905 |
0.54 |
| chr4_82613830_82614265 | 0.67 |
Gm11267 |
predicted gene 11267 |
26728 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.4 | 1.8 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.4 | 2.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 0.9 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.3 | 0.8 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.3 | 0.5 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 | 1.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.7 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 0.9 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.1 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.2 | 0.7 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 0.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.6 | GO:1903802 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.2 | 1.0 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.2 | 0.6 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.5 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.2 | 1.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.4 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.5 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 1.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.2 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0021855 | hypothalamus cell migration(GO:0021855) |
| 0.1 | 0.7 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.3 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.4 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.2 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.0 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.1 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.1 | 0.3 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 2.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.0 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 0.0 | 0.8 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0048819 | positive regulation of hair follicle maturation(GO:0048818) regulation of hair follicle maturation(GO:0048819) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.2 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.0 | GO:1904683 | regulation of metalloendopeptidase activity(GO:1904683) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:1904938 | dopaminergic neuron axon guidance(GO:0036514) planar cell polarity pathway involved in axon guidance(GO:1904938) |
| 0.0 | 0.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.1 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.1 | GO:0048103 | somatic stem cell division(GO:0048103) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.0 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 0.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 1.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.1 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 5.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 4.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 2.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.2 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.9 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 2.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 0.1 | GO:0001032 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0052825 | JUN kinase phosphatase activity(GO:0008579) phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0019198 | transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.0 | 0.0 | GO:0043919 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |