Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
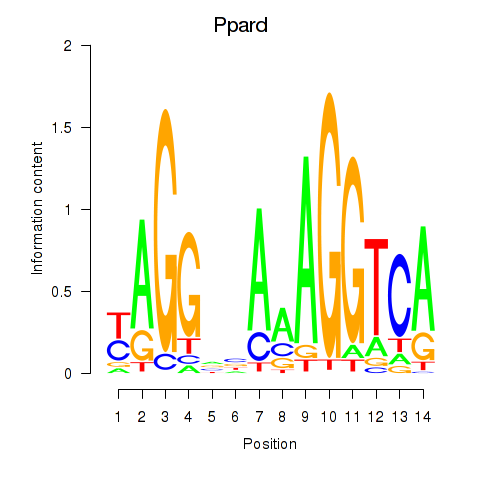
Results for Ppard
Z-value: 0.55
Transcription factors associated with Ppard
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppard
|
ENSMUSG00000002250.9 | peroxisome proliferator activator receptor delta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
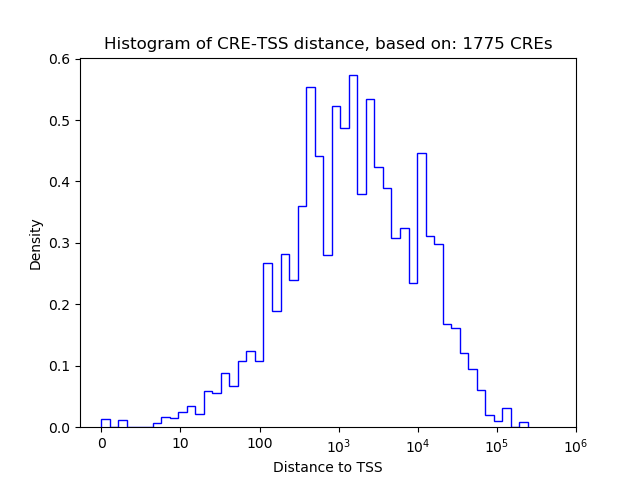
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_28237484_28238264 | Ppard | 5067 | 0.123219 | 0.30 | 2.0e-02 | Click! |
| chr17_28299783_28299934 | Ppard | 6612 | 0.105217 | -0.29 | 2.4e-02 | Click! |
| chr17_28237285_28237456 | Ppard | 4563 | 0.126914 | 0.28 | 3.1e-02 | Click! |
| chr17_28236269_28236726 | Ppard | 3690 | 0.137032 | 0.26 | 4.6e-02 | Click! |
| chr17_28236952_28237103 | Ppard | 4220 | 0.130181 | 0.24 | 6.4e-02 | Click! |
Activity of the Ppard motif across conditions
Conditions sorted by the z-value of the Ppard motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
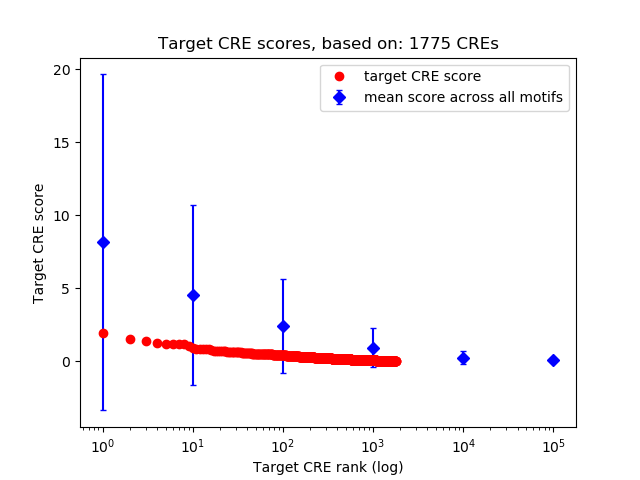
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_75655315_75655603 | 1.91 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
11274 |
0.18 |
| chr4_152447653_152448768 | 1.52 |
Kcnab2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
531 |
0.76 |
| chr15_78245409_78246081 | 1.39 |
Ncf4 |
neutrophil cytosolic factor 4 |
901 |
0.49 |
| chr11_103102696_103105788 | 1.27 |
Acbd4 |
acyl-Coenzyme A binding domain containing 4 |
463 |
0.7 |
| chr5_119685576_119687800 | 1.17 |
Tbx3os2 |
T-box 3, opposite strand 2 |
4530 |
0.17 |
| chr4_118429529_118430133 | 1.16 |
Elovl1 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
130 |
0.92 |
| chr6_56900814_56901268 | 1.16 |
Nt5c3 |
5'-nucleotidase, cytosolic III |
842 |
0.51 |
| chr1_91057148_91057757 | 1.16 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
3866 |
0.24 |
| chr2_167691999_167692442 | 1.08 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
1039 |
0.35 |
| chr6_55324306_55324538 | 0.94 |
Aqp1 |
aquaporin 1 |
12010 |
0.14 |
| chr6_125312691_125314238 | 0.83 |
Ltbr |
lymphotoxin B receptor |
199 |
0.88 |
| chr10_75939108_75939976 | 0.82 |
Gm867 |
predicted gene 867 |
1069 |
0.25 |
| chr6_72391190_72391463 | 0.82 |
Vamp8 |
vesicle-associated membrane protein 8 |
623 |
0.55 |
| chr18_38248552_38248782 | 0.82 |
1700086O06Rik |
RIKEN cDNA 1700086O06 gene |
1211 |
0.27 |
| chr10_12812573_12812894 | 0.81 |
Utrn |
utrophin |
816 |
0.64 |
| chr6_72392212_72392477 | 0.74 |
Vamp8 |
vesicle-associated membrane protein 8 |
1641 |
0.22 |
| chr9_107982030_107984233 | 0.72 |
Gm20661 |
predicted gene 20661 |
38 |
0.54 |
| chr2_155988856_155989174 | 0.72 |
Cep250 |
centrosomal protein 250 |
3100 |
0.14 |
| chr2_35610114_35610843 | 0.72 |
Dab2ip |
disabled 2 interacting protein |
11503 |
0.2 |
| chr16_90739931_90740383 | 0.72 |
Mrap |
melanocortin 2 receptor accessory protein |
1833 |
0.28 |
| chr6_29271003_29271699 | 0.71 |
Hilpda |
hypoxia inducible lipid droplet associated |
1137 |
0.37 |
| chr11_98446534_98448432 | 0.68 |
Grb7 |
growth factor receptor bound protein 7 |
415 |
0.68 |
| chr8_111573318_111574249 | 0.67 |
Znrf1 |
zinc and ring finger 1 |
11943 |
0.18 |
| chr9_40323395_40324376 | 0.66 |
1700110K17Rik |
RIKEN cDNA 1700110K17 gene |
459 |
0.71 |
| chr2_85060620_85061523 | 0.66 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
107 |
0.95 |
| chr17_33778675_33780469 | 0.65 |
Angptl4 |
angiopoietin-like 4 |
429 |
0.65 |
| chr3_121200717_121201242 | 0.65 |
Gm5710 |
predicted gene 5710 |
19882 |
0.13 |
| chr5_107874374_107875235 | 0.64 |
Evi5 |
ecotropic viral integration site 5 |
240 |
0.86 |
| chr14_63270328_63271232 | 0.63 |
Gata4 |
GATA binding protein 4 |
344 |
0.87 |
| chr11_95778707_95778988 | 0.62 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
17356 |
0.11 |
| chr17_80833726_80834127 | 0.62 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
62867 |
0.11 |
| chr15_82379480_82380754 | 0.61 |
Cyp2d22 |
cytochrome P450, family 2, subfamily d, polypeptide 22 |
126 |
0.88 |
| chr7_127014020_127014913 | 0.60 |
Mvp |
major vault protein |
84 |
0.88 |
| chr6_83247043_83247210 | 0.60 |
Slc4a5 |
solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
9751 |
0.12 |
| chr2_181241290_181242534 | 0.59 |
Helz2 |
helicase with zinc finger 2, transcriptional coactivator |
51 |
0.95 |
| chr16_44015370_44016774 | 0.58 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr10_69348031_69348525 | 0.58 |
Cdk1 |
cyclin-dependent kinase 1 |
2552 |
0.25 |
| chr11_119392018_119392570 | 0.57 |
Rnf213 |
ring finger protein 213 |
806 |
0.49 |
| chr11_102356812_102357313 | 0.56 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
3506 |
0.13 |
| chr19_55941741_55942415 | 0.56 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
43769 |
0.16 |
| chr11_65266805_65267363 | 0.55 |
Myocd |
myocardin |
2770 |
0.29 |
| chr15_76118017_76119354 | 0.55 |
Eppk1 |
epiplakin 1 |
1510 |
0.2 |
| chr4_59196867_59197419 | 0.55 |
Ugcg |
UDP-glucose ceramide glucosyltransferase |
7585 |
0.18 |
| chr11_79071094_79071622 | 0.54 |
Ksr1 |
kinase suppressor of ras 1 |
3128 |
0.28 |
| chr7_97454829_97454980 | 0.54 |
Kctd14 |
potassium channel tetramerisation domain containing 14 |
1682 |
0.29 |
| chr14_48538424_48539155 | 0.53 |
4930572G02Rik |
RIKEN cDNA 4930572G02 gene |
430 |
0.76 |
| chr6_142470696_142471396 | 0.53 |
Gys2 |
glycogen synthase 2 |
2063 |
0.3 |
| chr11_83849571_83850989 | 0.53 |
Hnf1b |
HNF1 homeobox B |
217 |
0.83 |
| chr1_171388990_171389691 | 0.53 |
Arhgap30 |
Rho GTPase activating protein 30 |
341 |
0.72 |
| chr8_79680146_79681210 | 0.52 |
Tpd52-ps |
tumor protein D52, pseudogene |
128 |
0.95 |
| chr15_67127618_67128557 | 0.52 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
936 |
0.68 |
| chr5_91849602_91849753 | 0.52 |
Gm5558 |
predicted gene 5558 |
42604 |
0.1 |
| chr5_124049658_124050948 | 0.52 |
Gm43661 |
predicted gene 43661 |
2049 |
0.18 |
| chr11_32296600_32297646 | 0.52 |
Hba-a2 |
hemoglobin alpha, adult chain 2 |
495 |
0.66 |
| chr2_127364227_127365175 | 0.52 |
Adra2b |
adrenergic receptor, alpha 2b |
1415 |
0.34 |
| chr9_103287942_103288250 | 0.52 |
1300017J02Rik |
RIKEN cDNA 1300017J02 gene |
128 |
0.96 |
| chr8_94964158_94964452 | 0.52 |
Gm10286 |
predicted gene 10286 |
5449 |
0.13 |
| chr11_55073341_55073492 | 0.51 |
Ccdc69 |
coiled-coil domain containing 69 |
4691 |
0.15 |
| chr15_103453834_103454841 | 0.51 |
Nckap1l |
NCK associated protein 1 like |
510 |
0.7 |
| chr11_96343236_96346574 | 0.51 |
Hoxb3 |
homeobox B3 |
1136 |
0.24 |
| chr10_70127197_70127431 | 0.50 |
Ccdc6 |
coiled-coil domain containing 6 |
30193 |
0.2 |
| chr11_32283784_32284776 | 0.50 |
Hba-a1 |
hemoglobin alpha, adult chain 1 |
469 |
0.66 |
| chr16_30407828_30408280 | 0.50 |
Atp13a3 |
ATPase type 13A3 |
2079 |
0.35 |
| chr2_72180043_72180526 | 0.49 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
591 |
0.76 |
| chr9_123124082_123124233 | 0.49 |
Exosc7 |
exosome component 7 |
2228 |
0.19 |
| chr4_135405096_135405380 | 0.49 |
Gm12990 |
predicted gene 12990 |
1582 |
0.22 |
| chr5_113136296_113136937 | 0.48 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
1370 |
0.27 |
| chr13_37799590_37800028 | 0.48 |
Gm46411 |
predicted gene, 46411 |
19 |
0.97 |
| chr11_120530321_120531885 | 0.48 |
Gcgr |
glucagon receptor |
329 |
0.46 |
| chr3_154192384_154192535 | 0.48 |
Slc44a5 |
solute carrier family 44, member 5 |
30023 |
0.16 |
| chr10_80347716_80349512 | 0.48 |
Adamtsl5 |
ADAMTS-like 5 |
202 |
0.82 |
| chr3_87767082_87768667 | 0.47 |
Pear1 |
platelet endothelial aggregation receptor 1 |
1061 |
0.43 |
| chr12_105757848_105758442 | 0.47 |
Ak7 |
adenylate kinase 7 |
12870 |
0.17 |
| chr5_108674592_108675616 | 0.47 |
Slc26a1 |
solute carrier family 26 (sulfate transporter), member 1 |
319 |
0.8 |
| chr8_122697299_122697450 | 0.47 |
Gm10612 |
predicted gene 10612 |
486 |
0.63 |
| chr5_89456826_89457321 | 0.47 |
Gc |
vitamin D binding protein |
825 |
0.68 |
| chr6_72388069_72388633 | 0.46 |
Vamp8 |
vesicle-associated membrane protein 8 |
1560 |
0.22 |
| chr11_94997783_94998270 | 0.46 |
Ppp1r9b |
protein phosphatase 1, regulatory subunit 9B |
1962 |
0.2 |
| chr14_65380585_65381143 | 0.46 |
Zfp395 |
zinc finger protein 395 |
5471 |
0.19 |
| chr2_38531311_38531917 | 0.46 |
Gm35808 |
predicted gene, 35808 |
4093 |
0.15 |
| chr4_43522717_43524714 | 0.46 |
Tpm2 |
tropomyosin 2, beta |
50 |
0.93 |
| chr11_20634522_20634924 | 0.46 |
Sertad2 |
SERTA domain containing 2 |
2744 |
0.3 |
| chr11_109860046_109860775 | 0.46 |
1700023C21Rik |
RIKEN cDNA 1700023C21 gene |
14532 |
0.18 |
| chr9_22135450_22135674 | 0.46 |
Acp5 |
acid phosphatase 5, tartrate resistant |
129 |
0.9 |
| chr10_7473717_7473913 | 0.45 |
Ulbp1 |
UL16 binding protein 1 |
177 |
0.95 |
| chr10_4611597_4612860 | 0.45 |
Esr1 |
estrogen receptor 1 (alpha) |
207 |
0.95 |
| chr16_32612719_32613020 | 0.45 |
Tfrc |
transferrin receptor |
2363 |
0.22 |
| chr15_77754021_77754265 | 0.45 |
Apol8 |
apolipoprotein L 8 |
1096 |
0.34 |
| chr9_108047652_108047803 | 0.45 |
Gmppb |
GDP-mannose pyrophosphorylase B |
1515 |
0.14 |
| chr9_31253741_31253909 | 0.45 |
Gm7244 |
predicted gene 7244 |
20996 |
0.14 |
| chr18_61035503_61036684 | 0.45 |
Cdx1 |
caudal type homeobox 1 |
106 |
0.95 |
| chr8_25848584_25848735 | 0.44 |
Kcnu1 |
potassium channel, subfamily U, member 1 |
964 |
0.33 |
| chr6_90324065_90325476 | 0.43 |
Chst13 |
carbohydrate sulfotransferase 13 |
415 |
0.73 |
| chr4_150220525_150221094 | 0.43 |
Gm13094 |
predicted gene 13094 |
7226 |
0.14 |
| chr11_69915606_69917001 | 0.43 |
Gps2 |
G protein pathway suppressor 2 |
262 |
0.72 |
| chr7_44482378_44484029 | 0.43 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
1314 |
0.19 |
| chr8_13202008_13202379 | 0.42 |
2810030D12Rik |
RIKEN cDNA 2810030D12 gene |
1373 |
0.23 |
| chr8_79372752_79373201 | 0.42 |
Smad1 |
SMAD family member 1 |
16298 |
0.17 |
| chr3_19958576_19959194 | 0.42 |
Cp |
ceruloplasmin |
1601 |
0.35 |
| chr11_120628124_120628346 | 0.41 |
Mafg |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
1913 |
0.1 |
| chr7_140835042_140835946 | 0.41 |
Urah |
urate (5-hydroxyiso-) hydrolase |
130 |
0.88 |
| chr6_131383705_131383961 | 0.41 |
Gm22362 |
predicted gene, 22362 |
3427 |
0.17 |
| chr3_86797094_86797252 | 0.41 |
Dclk2 |
doublecortin-like kinase 2 |
839 |
0.63 |
| chr2_27246009_27247328 | 0.41 |
Sardh |
sarcosine dehydrogenase |
180 |
0.94 |
| chr4_106462824_106463256 | 0.41 |
Pcsk9 |
proprotein convertase subtilisin/kexin type 9 |
1289 |
0.37 |
| chr2_173031954_173032465 | 0.41 |
Gm14453 |
predicted gene 14453 |
2371 |
0.21 |
| chr8_122321302_122321696 | 0.40 |
Zfpm1 |
zinc finger protein, multitype 1 |
12199 |
0.12 |
| chr11_4117645_4117796 | 0.40 |
Sec14l2 |
SEC14-like lipid binding 2 |
976 |
0.33 |
| chr19_5662426_5664007 | 0.39 |
Sipa1 |
signal-induced proliferation associated gene 1 |
404 |
0.63 |
| chr9_57639659_57641273 | 0.39 |
Csk |
c-src tyrosine kinase |
4643 |
0.13 |
| chr5_96919772_96919923 | 0.38 |
Gm8013 |
predicted gene 8013 |
1425 |
0.23 |
| chr1_74036048_74037430 | 0.38 |
Tns1 |
tensin 1 |
394 |
0.88 |
| chr7_132494201_132494423 | 0.38 |
Gm4587 |
predicted gene 4587 |
10654 |
0.18 |
| chr3_58681851_58682065 | 0.38 |
Siah2 |
siah E3 ubiquitin protein ligase 2 |
4079 |
0.15 |
| chr16_8746356_8746577 | 0.37 |
Usp7 |
ubiquitin specific peptidase 7 |
8046 |
0.16 |
| chr8_84908331_84909758 | 0.37 |
Dnase2a |
deoxyribonuclease II alpha |
313 |
0.72 |
| chr5_36621046_36621447 | 0.37 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
9 |
0.96 |
| chr13_37345719_37346245 | 0.37 |
Ly86 |
lymphocyte antigen 86 |
593 |
0.71 |
| chr4_137048694_137049053 | 0.37 |
Zbtb40 |
zinc finger and BTB domain containing 40 |
72 |
0.97 |
| chr5_143528879_143529030 | 0.37 |
Rac1 |
Rac family small GTPase 1 |
918 |
0.45 |
| chr6_40585006_40585430 | 0.37 |
Clec5a |
C-type lectin domain family 5, member a |
572 |
0.65 |
| chr4_83315034_83315192 | 0.37 |
Ttc39b |
tetratricopeptide repeat domain 39B |
9076 |
0.19 |
| chr10_116558082_116558233 | 0.37 |
Gm49344 |
predicted gene, 49344 |
8625 |
0.14 |
| chr11_95778990_95779141 | 0.37 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
17574 |
0.11 |
| chrX_73909796_73911465 | 0.36 |
Arhgap4 |
Rho GTPase activating protein 4 |
599 |
0.57 |
| chr15_36644507_36644990 | 0.36 |
Gm6704 |
predicted gene 6704 |
14879 |
0.12 |
| chr2_158145151_158146425 | 0.35 |
Tgm2 |
transglutaminase 2, C polypeptide |
578 |
0.71 |
| chr11_73326431_73327128 | 0.35 |
Aspa |
aspartoacylase |
28 |
0.95 |
| chr2_131497015_131497229 | 0.35 |
Smox |
spermine oxidase |
4858 |
0.2 |
| chr1_135133209_135134183 | 0.35 |
Ptpn7 |
protein tyrosine phosphatase, non-receptor type 7 |
387 |
0.67 |
| chr14_55105801_55106847 | 0.35 |
Ap1g2 |
adaptor protein complex AP-1, gamma 2 subunit |
57 |
0.94 |
| chr2_30266436_30267404 | 0.35 |
Phyhd1 |
phytanoyl-CoA dioxygenase domain containing 1 |
57 |
0.94 |
| chr11_73136220_73136518 | 0.35 |
Haspin |
histone H3 associated protein kinase |
1925 |
0.22 |
| chr2_180724979_180726144 | 0.35 |
Slc17a9 |
solute carrier family 17, member 9 |
161 |
0.92 |
| chr4_8707697_8708271 | 0.34 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
2356 |
0.38 |
| chr6_136938496_136939168 | 0.34 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
759 |
0.55 |
| chr11_98586074_98586564 | 0.34 |
Ormdl3 |
ORM1-like 3 (S. cerevisiae) |
1049 |
0.35 |
| chr10_59403301_59404650 | 0.34 |
Pla2g12b |
phospholipase A2, group XIIB |
315 |
0.88 |
| chr7_83880797_83881971 | 0.34 |
Tlnrd1 |
talin rod domain containing 1 |
2921 |
0.13 |
| chrX_53268548_53269792 | 0.34 |
Fam122b |
family with sequence similarity 122, member B |
150 |
0.94 |
| chr10_59440391_59440627 | 0.34 |
Oit3 |
oncoprotein induced transcript 3 |
1269 |
0.43 |
| chr2_173047915_173048486 | 0.34 |
Gm14453 |
predicted gene 14453 |
13620 |
0.13 |
| chr2_84936571_84938205 | 0.34 |
Slc43a3 |
solute carrier family 43, member 3 |
498 |
0.71 |
| chr17_81735907_81736099 | 0.34 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
2374 |
0.4 |
| chr4_11759825_11760000 | 0.34 |
Cdh17 |
cadherin 17 |
1719 |
0.41 |
| chr1_169453592_169453819 | 0.34 |
Gm5265 |
predicted pseudogene 5265 |
164 |
0.97 |
| chr3_88145024_88146544 | 0.34 |
Mef2d |
myocyte enhancer factor 2D |
3210 |
0.14 |
| chr15_89209951_89210279 | 0.34 |
Ppp6r2 |
protein phosphatase 6, regulatory subunit 2 |
1438 |
0.26 |
| chr4_57914999_57916744 | 0.33 |
D630039A03Rik |
RIKEN cDNA D630039A03 gene |
426 |
0.84 |
| chr7_109601527_109601678 | 0.33 |
Denn2b |
DENN domain containing 2B |
1063 |
0.5 |
| chr11_49050525_49051195 | 0.33 |
Olfr56 |
olfactory receptor 56 |
127 |
0.66 |
| chr11_4987942_4988093 | 0.33 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
1083 |
0.42 |
| chr12_76535555_76536098 | 0.32 |
Plekhg3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
2206 |
0.21 |
| chr6_87809573_87810010 | 0.32 |
Rab43 |
RAB43, member RAS oncogene family |
34 |
0.93 |
| chr4_117928946_117930077 | 0.32 |
Artn |
artemin |
14 |
0.96 |
| chr7_99237308_99238665 | 0.32 |
Mogat2 |
monoacylglycerol O-acyltransferase 2 |
608 |
0.62 |
| chr7_102565536_102565805 | 0.32 |
Trim21 |
tripartite motif-containing 21 |
184 |
0.84 |
| chr14_40992686_40992932 | 0.32 |
Prxl2a |
peroxiredoxin like 2A |
10295 |
0.17 |
| chr4_43037093_43037817 | 0.32 |
Fam214b |
family with sequence similarity 214, member B |
154 |
0.91 |
| chr3_59129062_59129246 | 0.32 |
P2ry14 |
purinergic receptor P2Y, G-protein coupled, 14 |
1468 |
0.38 |
| chr5_35727822_35729089 | 0.32 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
557 |
0.73 |
| chr12_109452854_109454456 | 0.32 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
0 |
0.96 |
| chr5_139793597_139793944 | 0.32 |
Mafk |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
2236 |
0.2 |
| chr4_107921827_107922539 | 0.32 |
Cpt2 |
carnitine palmitoyltransferase 2 |
1264 |
0.35 |
| chr10_78086930_78087081 | 0.31 |
Icosl |
icos ligand |
13666 |
0.12 |
| chr15_78412939_78413161 | 0.31 |
Mpst |
mercaptopyruvate sulfurtransferase |
3068 |
0.12 |
| chr9_62420746_62421699 | 0.31 |
Coro2b |
coronin, actin binding protein, 2B |
6779 |
0.24 |
| chr4_117840171_117840322 | 0.31 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
4808 |
0.13 |
| chr8_105850624_105850775 | 0.31 |
Cenpt |
centromere protein T |
1270 |
0.23 |
| chr13_73767696_73767973 | 0.31 |
Slc12a7 |
solute carrier family 12, member 7 |
4095 |
0.21 |
| chr7_110097583_110097734 | 0.31 |
Zfp143 |
zinc finger protein 143 |
6029 |
0.13 |
| chr9_42460796_42461392 | 0.31 |
Tbcel |
tubulin folding cofactor E-like |
367 |
0.85 |
| chr10_18446906_18447110 | 0.30 |
Nhsl1 |
NHS-like 1 |
22880 |
0.22 |
| chr15_83165887_83166710 | 0.30 |
Cyb5r3 |
cytochrome b5 reductase 3 |
3879 |
0.12 |
| chr19_53796222_53796373 | 0.30 |
Rbm20 |
RNA binding motif protein 20 |
2989 |
0.23 |
| chr11_75165245_75169157 | 0.30 |
Hic1 |
hypermethylated in cancer 1 |
945 |
0.35 |
| chr1_129956479_129957256 | 0.30 |
Gm37278 |
predicted gene, 37278 |
13705 |
0.22 |
| chr6_145121642_145121793 | 0.30 |
Lrmp |
lymphoid-restricted membrane protein |
22 |
0.97 |
| chr2_170153321_170154276 | 0.30 |
Zfp217 |
zinc finger protein 217 |
5695 |
0.31 |
| chr7_78914537_78914688 | 0.29 |
Isg20 |
interferon-stimulated protein |
283 |
0.86 |
| chr13_9107287_9107639 | 0.29 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
13481 |
0.15 |
| chr5_52990921_52991150 | 0.29 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
1257 |
0.4 |
| chr5_149265060_149265699 | 0.29 |
Alox5ap |
arachidonate 5-lipoxygenase activating protein |
32 |
0.95 |
| chrX_85577312_85577893 | 0.29 |
Tab3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
3452 |
0.27 |
| chr11_87748405_87749482 | 0.29 |
Mir142hg |
Mir142 host gene (non-protein coding) |
6634 |
0.09 |
| chr10_78297357_78298225 | 0.29 |
Agpat3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
407 |
0.49 |
| chr11_97442581_97443059 | 0.29 |
Arhgap23 |
Rho GTPase activating protein 23 |
6535 |
0.17 |
| chr4_133007332_133008511 | 0.29 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
3339 |
0.22 |
| chr14_48589666_48590768 | 0.28 |
Gm49120 |
predicted gene, 49120 |
382 |
0.79 |
| chr19_53186353_53186984 | 0.28 |
Add3 |
adducin 3 (gamma) |
5072 |
0.18 |
| chr5_122292013_122292461 | 0.28 |
Pptc7 |
PTC7 protein phosphatase homolog |
3535 |
0.14 |
| chr19_5724002_5724609 | 0.28 |
Ehbp1l1 |
EH domain binding protein 1-like 1 |
1965 |
0.11 |
| chr5_24855106_24855299 | 0.28 |
Rheb |
Ras homolog enriched in brain |
12578 |
0.14 |
| chr16_91929524_91929738 | 0.28 |
D430001F17Rik |
RIKEN cDNA D430001F17 gene |
1606 |
0.21 |
| chr8_117201307_117201734 | 0.28 |
Gan |
giant axonal neuropathy |
43383 |
0.14 |
| chr17_86293099_86293835 | 0.28 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
6289 |
0.29 |
| chr1_87573319_87573846 | 0.28 |
Ngef |
neuronal guanine nucleotide exchange factor |
288 |
0.62 |
| chr1_193001250_193001648 | 0.28 |
Syt14 |
synaptotagmin XIV |
34195 |
0.13 |
| chr7_128205575_128206445 | 0.28 |
Cox6a2 |
cytochrome c oxidase subunit 6A2 |
377 |
0.67 |
| chr18_48693060_48693350 | 0.27 |
Gm50229 |
predicted gene, 50229 |
29481 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.2 | 1.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.6 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.2 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.2 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.6 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.2 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.2 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.4 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0071655 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.0 | 0.0 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.0 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.6 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.2 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.0 | GO:0001781 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0006548 | histidine metabolic process(GO:0006547) histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.0 | 0.0 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.4 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0045404 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.0 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.0 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0035928 | RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.2 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.2 | GO:0044105 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.4 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.4 | GO:0043883 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.0 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0016675 | oxidoreductase activity, acting on a heme group of donors(GO:0016675) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |