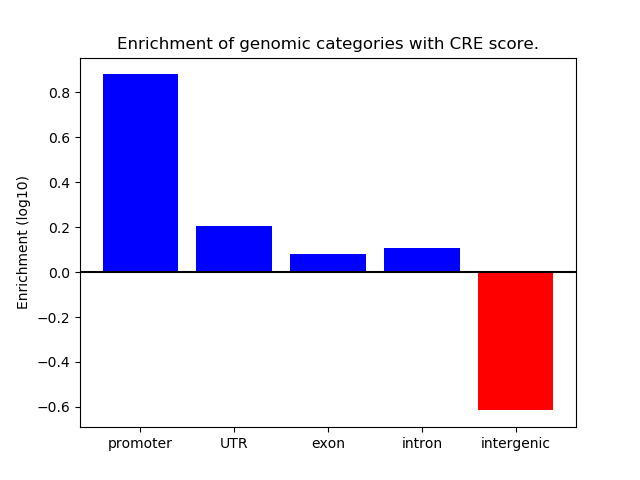
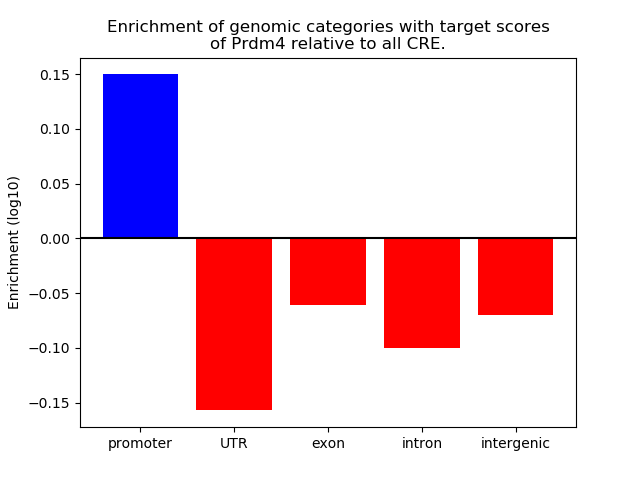
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
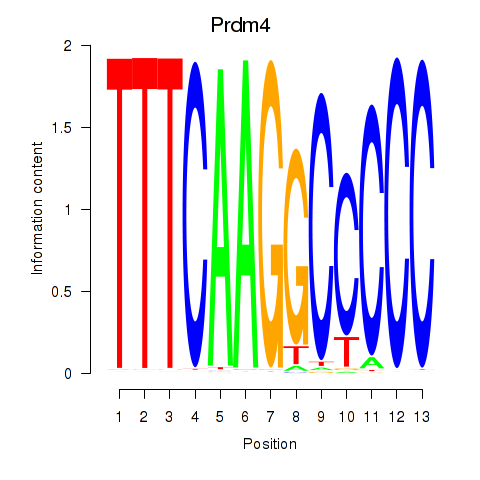
Results for Prdm4
Z-value: 0.37
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSMUSG00000035529.9 | PR domain containing 4 |
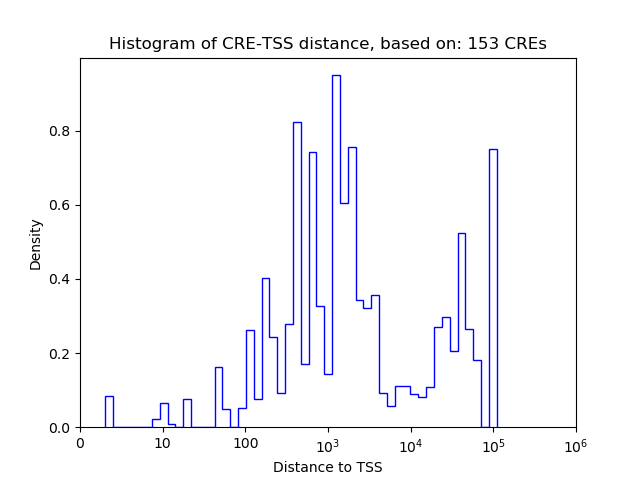
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_85917236_85917429 | Prdm4 | 190 | 0.906396 | 0.26 | 4.6e-02 | Click! |
| chr10_85914196_85914347 | Prdm4 | 2385 | 0.182928 | 0.11 | 4.0e-01 | Click! |
| chr10_85915840_85916954 | Prdm4 | 259 | 0.864648 | -0.01 | 9.5e-01 | Click! |
Activity of the Prdm4 motif across conditions
Conditions sorted by the z-value of the Prdm4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
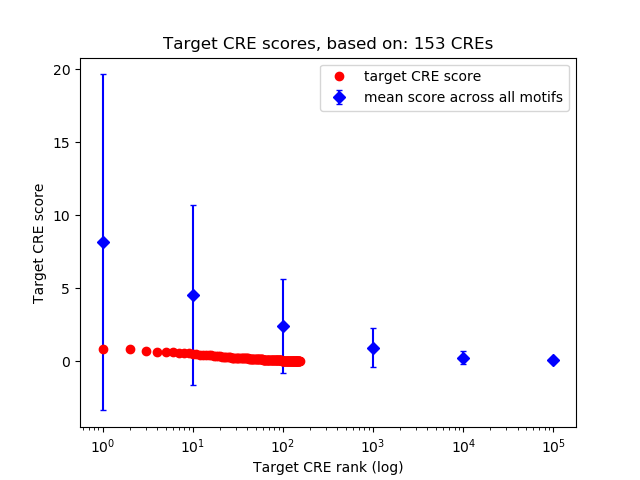
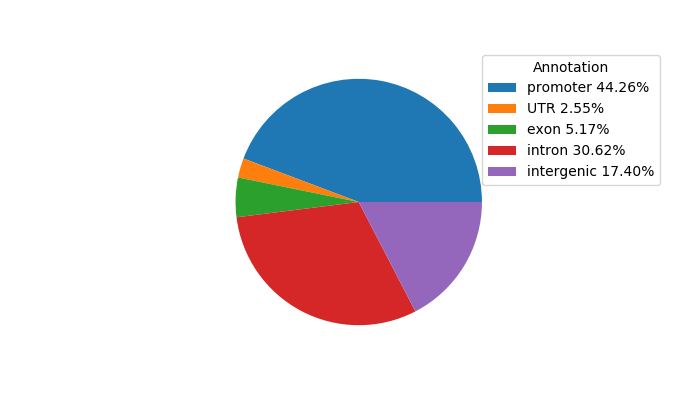
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_41710667_41710890 | 0.87 |
Oxr1 |
oxidation resistance 1 |
174 |
0.97 |
| chr11_63712018_63712440 | 0.86 |
Gm12288 |
predicted gene 12288 |
106071 |
0.07 |
| chr11_76353568_76353719 | 0.72 |
Nxn |
nucleoredoxin |
45425 |
0.12 |
| chr17_24473380_24473531 | 0.66 |
Bricd5 |
BRICHOS domain containing 5 |
429 |
0.59 |
| chr7_120173917_120175138 | 0.64 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
669 |
0.61 |
| chr14_119908730_119908881 | 0.64 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
109827 |
0.06 |
| chr2_74680662_74681213 | 0.58 |
Hoxd11 |
homeobox D11 |
1380 |
0.15 |
| chr2_154568646_154569144 | 0.57 |
E2f1 |
E2F transcription factor 1 |
825 |
0.45 |
| chr12_73476375_73476773 | 0.54 |
Gm48656 |
predicted gene, 48656 |
19667 |
0.14 |
| chr3_106545478_106545703 | 0.51 |
Cept1 |
choline/ethanolaminephosphotransferase 1 |
1992 |
0.2 |
| chr12_80111226_80112041 | 0.48 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
1361 |
0.31 |
| chr11_83849571_83850989 | 0.45 |
Hnf1b |
HNF1 homeobox B |
217 |
0.83 |
| chr11_32383101_32383252 | 0.44 |
Sh3pxd2b |
SH3 and PX domains 2B |
35336 |
0.13 |
| chr5_103428210_103428361 | 0.42 |
Ptpn13 |
protein tyrosine phosphatase, non-receptor type 13 |
2784 |
0.29 |
| chr10_87420108_87420324 | 0.41 |
Gm23191 |
predicted gene, 23191 |
56070 |
0.12 |
| chr8_121085538_121086180 | 0.40 |
Gm27530 |
predicted gene, 27530 |
1153 |
0.3 |
| chr4_98108504_98108946 | 0.38 |
Gm12691 |
predicted gene 12691 |
37874 |
0.2 |
| chr6_87993752_87994823 | 0.35 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
108 |
0.92 |
| chr15_73148296_73148447 | 0.35 |
Ago2 |
argonaute RISC catalytic subunit 2 |
29476 |
0.16 |
| chr7_142662290_142664788 | 0.34 |
Igf2os |
insulin-like growth factor 2, opposite strand |
1599 |
0.21 |
| chr2_172368075_172368471 | 0.32 |
Aurka |
aurora kinase A |
2233 |
0.19 |
| chr17_43671571_43671722 | 0.28 |
Cyp39a1 |
cytochrome P450, family 39, subfamily a, polypeptide 1 |
4121 |
0.18 |
| chr11_98922257_98922979 | 0.28 |
Cdc6 |
cell division cycle 6 |
2152 |
0.18 |
| chr3_81039797_81040176 | 0.27 |
Gm16000 |
predicted gene 16000 |
451 |
0.78 |
| chr13_63241575_63242090 | 0.26 |
Aopep |
aminopeptidase O |
1724 |
0.2 |
| chr6_28425695_28425934 | 0.26 |
Gcc1 |
golgi coiled coil 1 |
1661 |
0.21 |
| chr8_84200619_84201949 | 0.26 |
Gm37352 |
predicted gene, 37352 |
415 |
0.58 |
| chr1_172310298_172310642 | 0.26 |
Igsf8 |
immunoglobulin superfamily, member 8 |
1301 |
0.28 |
| chr15_89185418_89185569 | 0.25 |
Mir6958 |
microRNA 6958 |
44 |
0.94 |
| chr2_105132016_105133665 | 0.24 |
Wt1 |
Wilms tumor 1 homolog |
1957 |
0.31 |
| chr11_96872293_96874152 | 0.23 |
Gm11523 |
predicted gene 11523 |
712 |
0.46 |
| chr17_53482249_53482404 | 0.22 |
Rab5a |
RAB5A, member RAS oncogene family |
3039 |
0.19 |
| chr8_45913620_45913830 | 0.22 |
Pdlim3 |
PDZ and LIM domain 3 |
1307 |
0.32 |
| chr2_32539078_32539941 | 0.22 |
Fam102a |
family with sequence similarity 102, member A |
681 |
0.5 |
| chr13_35473350_35473501 | 0.21 |
Gm48704 |
predicted gene, 48704 |
70477 |
0.1 |
| chr3_129201144_129201586 | 0.21 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
1396 |
0.39 |
| chr19_60146143_60147364 | 0.20 |
E330013P04Rik |
RIKEN cDNA E330013P04 gene |
2049 |
0.34 |
| chr3_89526596_89526945 | 0.20 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
6606 |
0.16 |
| chr11_94939289_94939965 | 0.20 |
Col1a1 |
collagen, type I, alpha 1 |
615 |
0.61 |
| chr10_79945094_79946752 | 0.19 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
356 |
0.66 |
| chr14_73049116_73049339 | 0.19 |
Cysltr2 |
cysteinyl leukotriene receptor 2 |
113 |
0.97 |
| chr11_99435806_99436036 | 0.18 |
Krt20 |
keratin 20 |
2229 |
0.16 |
| chr15_103058659_103059955 | 0.18 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
1 |
0.95 |
| chr10_93309488_93311306 | 0.18 |
Elk3 |
ELK3, member of ETS oncogene family |
414 |
0.82 |
| chr7_45174584_45175709 | 0.17 |
Mir7055 |
microRNA 7055 |
1843 |
0.1 |
| chr15_79040318_79041315 | 0.17 |
Galr3 |
galanin receptor 3 |
1069 |
0.25 |
| chr3_99261050_99262399 | 0.17 |
Gm43120 |
predicted gene 43120 |
439 |
0.81 |
| chr3_95011750_95011901 | 0.16 |
Zfp687 |
zinc finger protein 687 |
18 |
0.94 |
| chr1_180943258_180943409 | 0.16 |
Tmem63a |
transmembrane protein 63a |
702 |
0.5 |
| chr11_95516491_95516808 | 0.16 |
Nxph3 |
neurexophilin 3 |
2079 |
0.28 |
| chr19_7153054_7153338 | 0.16 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
47268 |
0.08 |
| chr17_28348327_28348971 | 0.16 |
Tead3 |
TEA domain family member 3 |
1697 |
0.21 |
| chrX_64747210_64747361 | 0.15 |
Gm14676 |
predicted gene 14676 |
3973 |
0.31 |
| chr5_144965930_144966401 | 0.15 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
318 |
0.86 |
| chr17_25415111_25416091 | 0.15 |
Cacna1h |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
17677 |
0.1 |
| chr3_55517135_55518024 | 0.14 |
Gm42608 |
predicted gene 42608 |
10 |
0.97 |
| chr8_110503112_110503371 | 0.13 |
Gm47306 |
predicted gene, 47306 |
4256 |
0.24 |
| chr19_55253303_55254683 | 0.13 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
624 |
0.71 |
| chr18_14063182_14063853 | 0.12 |
Zfp521 |
zinc finger protein 521 |
90730 |
0.09 |
| chr13_63258769_63259476 | 0.12 |
Gm47603 |
predicted gene, 47603 |
3558 |
0.11 |
| chr8_36745950_36746830 | 0.12 |
Dlc1 |
deleted in liver cancer 1 |
13336 |
0.28 |
| chr13_55182504_55182904 | 0.11 |
Fgfr4 |
fibroblast growth factor receptor 4 |
26587 |
0.11 |
| chr14_72600187_72600734 | 0.11 |
Fndc3a |
fibronectin type III domain containing 3A |
2502 |
0.3 |
| chr1_72873610_72874834 | 0.11 |
Igfbp5 |
insulin-like growth factor binding protein 5 |
53 |
0.98 |
| chr12_26454705_26455291 | 0.10 |
Rsad2 |
radical S-adenosyl methionine domain containing 2 |
156 |
0.94 |
| chr13_48977850_48978331 | 0.10 |
Fam120a |
family with sequence similarity 120, member A |
10073 |
0.24 |
| chr8_121396512_121396947 | 0.10 |
Gm26747 |
predicted gene, 26747 |
24509 |
0.15 |
| chr3_105914133_105914707 | 0.10 |
Tmigd3 |
transmembrane and immunoglobulin domain containing 3 |
284 |
0.84 |
| chr5_17584554_17584705 | 0.09 |
Sema3c |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
9730 |
0.28 |
| chr17_43025342_43025543 | 0.09 |
Tnfrsf21 |
tumor necrosis factor receptor superfamily, member 21 |
8874 |
0.29 |
| chr18_42053706_42054633 | 0.09 |
Sh3rf2 |
SH3 domain containing ring finger 2 |
459 |
0.84 |
| chr7_19563781_19564198 | 0.08 |
Ppp1r37 |
protein phosphatase 1, regulatory subunit 37 |
913 |
0.36 |
| chr12_81816545_81817321 | 0.08 |
Gm47080 |
predicted gene, 47080 |
10866 |
0.17 |
| chr10_81280318_81280649 | 0.08 |
Tjp3 |
tight junction protein 3 |
366 |
0.64 |
| chr17_56461082_56462187 | 0.08 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
527 |
0.73 |
| chr13_3891572_3893506 | 0.08 |
Net1 |
neuroepithelial cell transforming gene 1 |
894 |
0.46 |
| chr18_12120723_12121070 | 0.08 |
Tmem241 |
transmembrane protein 241 |
572 |
0.72 |
| chr1_66700897_66701735 | 0.08 |
Rpe |
ribulose-5-phosphate-3-epimerase |
409 |
0.78 |
| chr17_80825543_80826587 | 0.08 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
70728 |
0.1 |
| chr17_68000612_68000868 | 0.07 |
Arhgap28 |
Rho GTPase activating protein 28 |
3380 |
0.35 |
| chr7_30584453_30584699 | 0.07 |
Kmt2b |
lysine (K)-specific methyltransferase 2B |
498 |
0.48 |
| chr17_32319078_32319484 | 0.07 |
Akap8 |
A kinase (PRKA) anchor protein 8 |
477 |
0.71 |
| chr1_161070981_161071821 | 0.07 |
Cenpl |
centromere protein L |
385 |
0.54 |
| chr4_139829324_139829858 | 0.07 |
Pax7 |
paired box 7 |
3416 |
0.28 |
| chr9_40325276_40326082 | 0.06 |
1700110K17Rik |
RIKEN cDNA 1700110K17 gene |
2253 |
0.19 |
| chr3_149093055_149093671 | 0.06 |
Gm25127 |
predicted gene, 25127 |
64431 |
0.11 |
| chr7_24904441_24905054 | 0.06 |
Arhgef1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
307 |
0.78 |
| chr2_69612307_69612590 | 0.06 |
Lrp2 |
low density lipoprotein receptor-related protein 2 |
26383 |
0.15 |
| chr3_129881133_129881711 | 0.06 |
Pla2g12a |
phospholipase A2, group XIIA |
238 |
0.9 |
| chr7_89903398_89904159 | 0.06 |
Ccdc81 |
coiled-coil domain containing 81 |
149 |
0.95 |
| chr4_62406680_62406837 | 0.06 |
Cdc26 |
cell division cycle 26 |
1244 |
0.31 |
| chr13_21996613_21997784 | 0.06 |
Prss16 |
protease, serine 16 (thymus) |
8024 |
0.06 |
| chr11_70012013_70013226 | 0.06 |
Acadvl |
acyl-Coenzyme A dehydrogenase, very long chain |
45 |
0.86 |
| chr12_91683421_91686419 | 0.06 |
Gm16876 |
predicted gene, 16876 |
255 |
0.86 |
| chr17_33557999_33558350 | 0.06 |
Myo1f |
myosin IF |
2409 |
0.22 |
| chr11_25288195_25288807 | 0.06 |
Gm12069 |
predicted gene 12069 |
12762 |
0.23 |
| chr16_85132591_85133705 | 0.05 |
Gm49226 |
predicted gene, 49226 |
15681 |
0.18 |
| chr15_102971199_102971716 | 0.05 |
Mir196a-2 |
microRNA 196a-2 |
1893 |
0.15 |
| chr17_34646272_34646494 | 0.05 |
Atf6b |
activating transcription factor 6 beta |
763 |
0.3 |
| chr4_11703848_11705053 | 0.05 |
Gem |
GTP binding protein (gene overexpressed in skeletal muscle) |
7 |
0.98 |
| chr14_99303099_99303560 | 0.05 |
Klf5 |
Kruppel-like factor 5 |
4175 |
0.19 |
| chr12_80113642_80114602 | 0.05 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
1109 |
0.34 |
| chr11_74900076_74900873 | 0.05 |
Tsr1 |
TSR1 20S rRNA accumulation |
1239 |
0.27 |
| chr2_102900531_102901984 | 0.04 |
Cd44 |
CD44 antigen |
89 |
0.97 |
| chr11_70525408_70526199 | 0.04 |
Psmb6 |
proteasome (prosome, macropain) subunit, beta type 6 |
307 |
0.72 |
| chr12_51377639_51378186 | 0.04 |
Scfd1 |
Sec1 family domain containing 1 |
320 |
0.91 |
| chr5_122146089_122146622 | 0.04 |
Ccdc63 |
coiled-coil domain containing 63 |
5532 |
0.14 |
| chr8_88523833_88524755 | 0.04 |
Nkd1 |
naked cuticle 1 |
2940 |
0.29 |
| chrX_48053949_48055179 | 0.04 |
Apln |
apelin |
19711 |
0.2 |
| chr17_45279982_45280133 | 0.04 |
Gm24979 |
predicted gene, 24979 |
70091 |
0.1 |
| chrX_64781879_64782030 | 0.04 |
Gm26111 |
predicted gene, 26111 |
18557 |
0.21 |
| chr19_43618352_43618699 | 0.04 |
Gm22646 |
predicted gene, 22646 |
3921 |
0.13 |
| chr14_27044544_27045365 | 0.04 |
Il17rd |
interleukin 17 receptor D |
5698 |
0.22 |
| chr6_145855324_145856436 | 0.03 |
Gm43909 |
predicted gene, 43909 |
7417 |
0.17 |
| chr19_3718944_3720017 | 0.03 |
Gm36787 |
predicted gene, 36787 |
93 |
0.93 |
| chr6_135785665_135785816 | 0.03 |
Gm22892 |
predicted gene, 22892 |
44743 |
0.15 |
| chr16_13903228_13903614 | 0.03 |
Mpv17l |
Mpv17 transgene, kidney disease mutant-like |
260 |
0.51 |
| chr11_87853248_87853905 | 0.03 |
Mks1 |
Meckel syndrome, type 1 |
89 |
0.94 |
| chr11_18750877_18751932 | 0.03 |
Gm28401 |
predicted gene 28401 |
5334 |
0.19 |
| chr12_110447124_110447282 | 0.03 |
Ppp2r5c |
protein phosphatase 2, regulatory subunit B', gamma |
49 |
0.97 |
| chr10_128547971_128549076 | 0.03 |
Rpl41 |
ribosomal protein L41 |
579 |
0.36 |
| chr18_25715120_25715657 | 0.03 |
0710001A04Rik |
RIKEN cDNA 0710001A04 gene |
1618 |
0.46 |
| chr19_43729441_43730557 | 0.03 |
Entpd7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
3632 |
0.16 |
| chr4_127025301_127025452 | 0.02 |
Sfpq |
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
548 |
0.64 |
| chr9_14717687_14717910 | 0.02 |
Piwil4 |
piwi-like RNA-mediated gene silencing 4 |
675 |
0.51 |
| chr5_128703817_128704666 | 0.02 |
Piwil1 |
piwi-like RNA-mediated gene silencing 1 |
1717 |
0.34 |
| chr10_85102050_85102712 | 0.02 |
Fhl4 |
four and a half LIM domains 4 |
114 |
0.68 |
| chr17_57249307_57250786 | 0.02 |
Trip10 |
thyroid hormone receptor interactor 10 |
498 |
0.63 |
| chr3_116807540_116808184 | 0.02 |
Agl |
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
11 |
0.96 |
| chr11_75291131_75291523 | 0.02 |
Gm47300 |
predicted gene, 47300 |
27281 |
0.12 |
| chr14_45332844_45332995 | 0.02 |
Psmc6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
1488 |
0.29 |
| chr2_80315446_80316199 | 0.02 |
Dnajc10 |
DnaJ heat shock protein family (Hsp40) member C10 |
356 |
0.89 |
| chr19_5057787_5057938 | 0.02 |
Rin1 |
Ras and Rab interactor 1 |
4757 |
0.07 |
| chr2_39004499_39005146 | 0.01 |
Rpl35 |
ribosomal protein L35 |
221 |
0.72 |
| chr3_55347442_55348146 | 0.01 |
Dclk1 |
doublecortin-like kinase 1 |
6340 |
0.18 |
| chr16_87454687_87455921 | 0.01 |
Usp16 |
ubiquitin specific peptidase 16 |
246 |
0.89 |
| chr2_92374659_92375167 | 0.01 |
Large2 |
LARGE xylosyl- and glucuronyltransferase 2 |
44 |
0.74 |
| chr3_9605900_9606819 | 0.01 |
Zfp704 |
zinc finger protein 704 |
3726 |
0.28 |
| chr7_135539645_135539955 | 0.01 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
1961 |
0.3 |
| chr4_41506381_41507061 | 0.01 |
1110017D15Rik |
RIKEN cDNA 1110017D15 gene |
1033 |
0.35 |
| chr5_113858981_113859268 | 0.01 |
Coro1c |
coronin, actin binding protein 1C |
350 |
0.71 |
| chr9_65169040_65169256 | 0.01 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
11590 |
0.13 |
| chr18_61637854_61638670 | 0.01 |
Bvht |
braveheart long non-coding RNA |
1280 |
0.31 |
| chrX_95163813_95164963 | 0.00 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
1437 |
0.48 |
| chr14_16597821_16598428 | 0.00 |
Rarb |
retinoic acid receptor, beta |
22297 |
0.21 |
| chr7_127009057_127009208 | 0.00 |
Mvp |
major vault protein |
2733 |
0.08 |
| chr3_89451824_89451975 | 0.00 |
Pmvk |
phosphomevalonate kinase |
2642 |
0.13 |
| chrX_109094919_109095373 | 0.00 |
Sh3bgrl |
SH3-binding domain glutamic acid-rich protein like |
219 |
0.94 |
| chr17_23630249_23630847 | 0.00 |
Gm49935 |
predicted gene, 49935 |
6048 |
0.07 |
| chr14_66911778_66912249 | 0.00 |
Pnma2 |
paraneoplastic antigen MA2 |
771 |
0.58 |
| chr14_34373115_34373266 | 0.00 |
Sncg |
synuclein, gamma |
1599 |
0.23 |
| chr6_141249439_141250607 | 0.00 |
Gm28523 |
predicted gene 28523 |
14 |
0.53 |
| chr8_57317473_57317624 | 0.00 |
Hand2os1 |
Hand2, opposite strand 1 |
1276 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.4 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) deltoid tuberosity development(GO:0035993) left lung morphogenesis(GO:0060460) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:1900623 | positive regulation of neutrophil apoptotic process(GO:0033031) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0050883 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |