Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
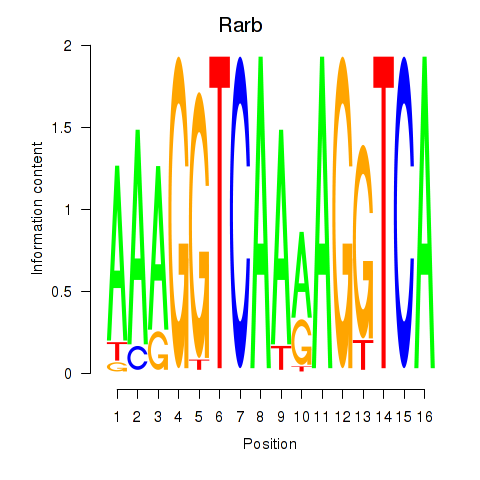
Results for Rarb
Z-value: 0.52
Transcription factors associated with Rarb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarb
|
ENSMUSG00000017491.8 | retinoic acid receptor, beta |
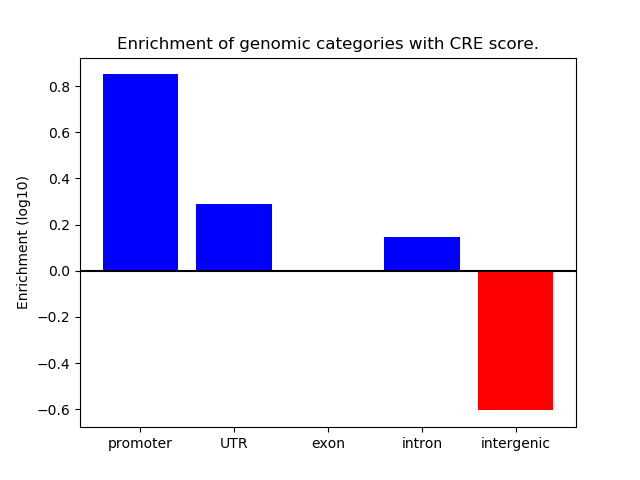
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_16574044_16575901 | Rarb | 73 | 0.919664 | -0.43 | 6.8e-04 | Click! |
| chr14_16554767_16555445 | Rarb | 19939 | 0.206185 | 0.33 | 1.0e-02 | Click! |
| chr14_16554164_16554315 | Rarb | 20806 | 0.204451 | 0.32 | 1.1e-02 | Click! |
| chr14_16847192_16847623 | Rarb | 28251 | 0.227952 | -0.31 | 1.6e-02 | Click! |
| chr14_16733832_16734108 | Rarb | 85035 | 0.097670 | 0.21 | 1.0e-01 | Click! |
Activity of the Rarb motif across conditions
Conditions sorted by the z-value of the Rarb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
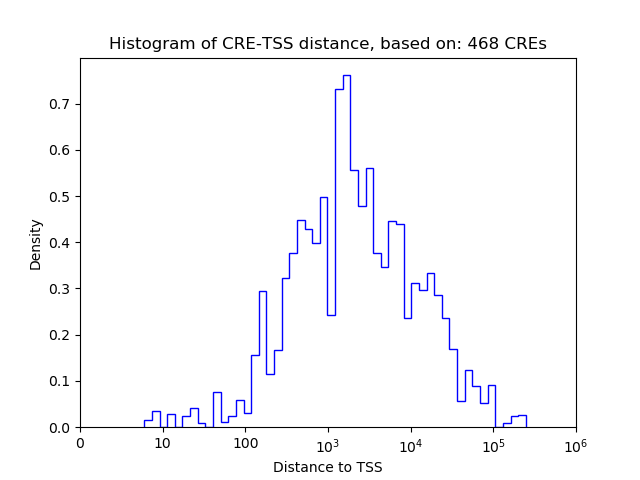
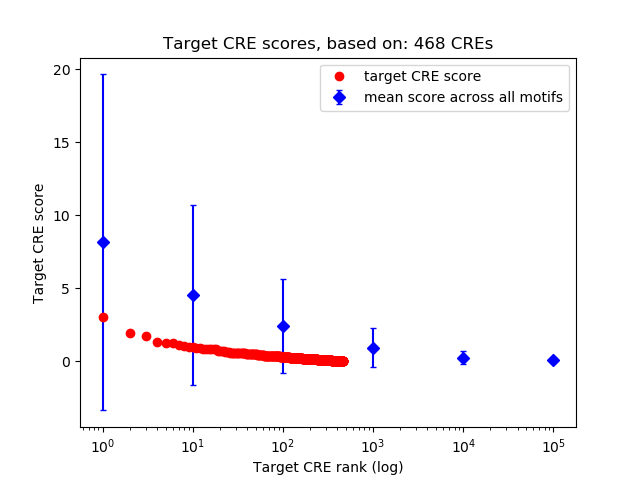
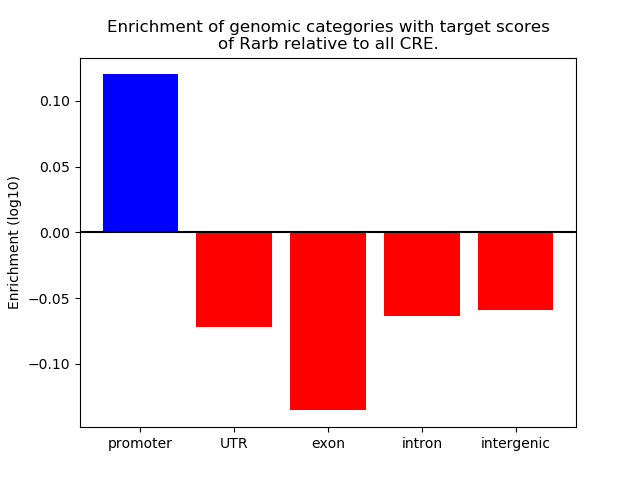
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_102360845_102363484 | 3.04 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
1540 |
0.24 |
| chr8_23035959_23037041 | 1.91 |
Ank1 |
ankyrin 1, erythroid |
1269 |
0.45 |
| chrX_150547515_150548479 | 1.76 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
538 |
0.44 |
| chr9_121451719_121452290 | 1.30 |
Trak1 |
trafficking protein, kinesin binding 1 |
2292 |
0.28 |
| chr11_58275551_58275913 | 1.27 |
4930438A08Rik |
RIKEN cDNA 4930438A08 gene |
672 |
0.56 |
| chr6_83068298_83071797 | 1.23 |
Tlx2 |
T cell leukemia, homeobox 2 |
178 |
0.81 |
| chr1_177494861_177495402 | 1.10 |
Gm37306 |
predicted gene, 37306 |
27753 |
0.14 |
| chr2_173034791_173035156 | 1.06 |
Gm14453 |
predicted gene 14453 |
393 |
0.8 |
| chr11_83848300_83848853 | 0.98 |
Gm12576 |
predicted gene 12576 |
917 |
0.43 |
| chr8_67948178_67948899 | 0.97 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
3341 |
0.28 |
| chr11_19986765_19987519 | 0.91 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
32572 |
0.2 |
| chr1_157526787_157526938 | 0.89 |
Sec16b |
SEC16 homolog B (S. cerevisiae) |
715 |
0.64 |
| chr10_77109606_77109767 | 0.87 |
Col18a1 |
collagen, type XVIII, alpha 1 |
4019 |
0.21 |
| chr3_131269218_131269951 | 0.87 |
Hadh |
hydroxyacyl-Coenzyme A dehydrogenase |
2440 |
0.24 |
| chr11_95354322_95355144 | 0.85 |
Fam117a |
family with sequence similarity 117, member A |
14771 |
0.11 |
| chr15_35295781_35297332 | 0.85 |
Osr2 |
odd-skipped related 2 |
445 |
0.83 |
| chr4_58546126_58546553 | 0.85 |
Lpar1 |
lysophosphatidic acid receptor 1 |
2062 |
0.34 |
| chr8_3354301_3354483 | 0.84 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
932 |
0.59 |
| chr1_172548699_172548872 | 0.72 |
4933439K11Rik |
RIKEN cDNA 4933439K11 gene |
1884 |
0.2 |
| chr5_52990921_52991150 | 0.72 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
1257 |
0.4 |
| chr14_21078036_21078470 | 0.72 |
Adk |
adenosine kinase |
2101 |
0.35 |
| chrX_106839224_106840351 | 0.71 |
Rtl3 |
retrotransposon Gag like 3 |
857 |
0.6 |
| chr15_77973908_77974338 | 0.66 |
Eif3d |
eukaryotic translation initiation factor 3, subunit D |
3310 |
0.19 |
| chr10_68157085_68157396 | 0.63 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
20614 |
0.24 |
| chr16_14310141_14310292 | 0.60 |
Gm15868 |
predicted gene 15868 |
1625 |
0.3 |
| chr17_65606813_65607080 | 0.60 |
Vapa |
vesicle-associated membrane protein, associated protein A |
6609 |
0.17 |
| chr10_86024993_86025483 | 0.60 |
A230060F14Rik |
RIKEN cDNA A230060F14 gene |
2909 |
0.15 |
| chr10_121263429_121264335 | 0.59 |
Gm35404 |
predicted gene, 35404 |
20880 |
0.14 |
| chr6_118771100_118771251 | 0.57 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
9149 |
0.27 |
| chr3_14641184_14642360 | 0.57 |
Car13 |
carbonic anhydrase 13 |
45 |
0.97 |
| chr8_70490321_70491265 | 0.57 |
Crlf1 |
cytokine receptor-like factor 1 |
2365 |
0.12 |
| chr4_109439636_109440032 | 0.56 |
Ttc39a |
tetratricopeptide repeat domain 39A |
18892 |
0.15 |
| chr10_8944239_8945864 | 0.55 |
Gm48728 |
predicted gene, 48728 |
6898 |
0.2 |
| chr7_25379357_25380698 | 0.55 |
4732471J01Rik |
RIKEN cDNA 4732471J01 gene |
3204 |
0.12 |
| chr9_22135450_22135674 | 0.54 |
Acp5 |
acid phosphatase 5, tartrate resistant |
129 |
0.9 |
| chr6_34517984_34518174 | 0.54 |
Gm13860 |
predicted gene 13860 |
15474 |
0.17 |
| chrX_38575133_38575425 | 0.54 |
Cul4b |
cullin 4B |
904 |
0.58 |
| chr3_95652966_95653117 | 0.53 |
Mcl1 |
myeloid cell leukemia sequence 1 |
5747 |
0.11 |
| chr5_137569837_137570642 | 0.53 |
Tfr2 |
transferrin receptor 2 |
370 |
0.67 |
| chr2_172261076_172261248 | 0.53 |
Mc3r |
melanocortin 3 receptor |
12670 |
0.17 |
| chr5_115945354_115946075 | 0.52 |
Cit |
citron |
417 |
0.82 |
| chr11_60477952_60478643 | 0.50 |
Myo15 |
myosin XV |
1618 |
0.23 |
| chr16_31066399_31067041 | 0.50 |
Xxylt1 |
xyloside xylosyltransferase 1 |
1133 |
0.51 |
| chr7_100463756_100463979 | 0.50 |
C2cd3 |
C2 calcium-dependent domain containing 3 |
344 |
0.77 |
| chr4_141558647_141558798 | 0.49 |
B330016D10Rik |
RIKEN cDNA B330016D10 gene |
12533 |
0.12 |
| chr1_149960555_149961570 | 0.49 |
Pla2g4a |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
193 |
0.96 |
| chr16_18286012_18286942 | 0.48 |
Gm24572 |
predicted gene, 24572 |
1840 |
0.14 |
| chr15_81870008_81870701 | 0.48 |
Phf5a |
PHD finger protein 5A |
1311 |
0.23 |
| chr11_53422380_53423106 | 0.48 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
427 |
0.66 |
| chr9_106357304_106357455 | 0.47 |
Dusp7 |
dual specificity phosphatase 7 |
11253 |
0.12 |
| chr2_116020004_116021542 | 0.47 |
Meis2 |
Meis homeobox 2 |
21273 |
0.19 |
| chr10_88349909_88350060 | 0.46 |
Dram1 |
DNA-damage regulated autophagy modulator 1 |
7012 |
0.17 |
| chr6_5287612_5288353 | 0.46 |
Pon2 |
paraoxonase 2 |
1086 |
0.49 |
| chr17_28007198_28009699 | 0.46 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
1103 |
0.37 |
| chr14_70597977_70598489 | 0.44 |
Fam160b2 |
family with sequence similarity 160, member B2 |
1602 |
0.25 |
| chr5_137481813_137482289 | 0.43 |
Epo |
erythropoietin |
3765 |
0.1 |
| chr6_52233543_52234485 | 0.43 |
Hoxa10 |
homeobox A10 |
690 |
0.34 |
| chr5_124574763_124574914 | 0.43 |
Eif2b1 |
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
78 |
0.94 |
| chr13_52283527_52283880 | 0.42 |
Gm48199 |
predicted gene, 48199 |
103292 |
0.07 |
| chr6_114873953_114875469 | 0.41 |
Vgll4 |
vestigial like family member 4 |
309 |
0.91 |
| chr8_105319792_105320155 | 0.40 |
Lrrc29 |
leucine rich repeat containing 29 |
6286 |
0.06 |
| chr1_75137178_75137539 | 0.39 |
Cnppd1 |
cyclin Pas1/PHO80 domain containing 1 |
448 |
0.64 |
| chr11_116429312_116429574 | 0.39 |
Ubald2 |
UBA-like domain containing 2 |
4651 |
0.12 |
| chr2_27981386_27981740 | 0.39 |
Col5a1 |
collagen, type V, alpha 1 |
35878 |
0.15 |
| chr17_12915129_12915280 | 0.39 |
Mrpl18 |
mitochondrial ribosomal protein L18 |
420 |
0.44 |
| chr4_124695736_124696563 | 0.39 |
Gm24480 |
predicted gene, 24480 |
1458 |
0.2 |
| chr11_107471149_107471443 | 0.39 |
Pitpnc1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
597 |
0.63 |
| chr1_193200286_193201469 | 0.38 |
Traf3ip3 |
TRAF3 interacting protein 3 |
457 |
0.66 |
| chr5_111336064_111336362 | 0.38 |
Pitpnb |
phosphatidylinositol transfer protein, beta |
5406 |
0.19 |
| chr5_24425282_24425881 | 0.38 |
Slc4a2 |
solute carrier family 4 (anion exchanger), member 2 |
296 |
0.73 |
| chr7_16230037_16230216 | 0.38 |
Gm45409 |
predicted gene 45409 |
1226 |
0.28 |
| chr14_33446117_33447122 | 0.38 |
Mapk8 |
mitogen-activated protein kinase 8 |
523 |
0.73 |
| chr7_116031040_116032871 | 0.38 |
Sox6os |
SRY (sex determining region Y)-box 6, opposite strand |
24 |
0.94 |
| chr7_49764281_49764691 | 0.37 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
5333 |
0.23 |
| chr10_45229498_45229649 | 0.37 |
Gm27723 |
predicted gene, 27723 |
17514 |
0.16 |
| chr5_130223795_130224186 | 0.37 |
Gm6598 |
predicted gene 6598 |
925 |
0.33 |
| chr15_12937941_12938422 | 0.37 |
Gm49100 |
predicted gene, 49100 |
17541 |
0.19 |
| chr19_4110129_4110280 | 0.37 |
Pitpnm1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
277 |
0.73 |
| chr6_72392212_72392477 | 0.36 |
Vamp8 |
vesicle-associated membrane protein 8 |
1641 |
0.22 |
| chr11_94997390_94997735 | 0.36 |
Ppp1r9b |
protein phosphatase 1, regulatory subunit 9B |
1498 |
0.25 |
| chr2_84843620_84843993 | 0.36 |
Slc43a1 |
solute carrier family 43, member 1 |
3181 |
0.15 |
| chr8_34061456_34061607 | 0.35 |
Gm9951 |
predicted gene 9951 |
6909 |
0.14 |
| chr4_154926952_154928851 | 0.35 |
Tnfrsf14 |
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
176 |
0.92 |
| chr2_84938213_84938457 | 0.35 |
Slc43a3 |
solute carrier family 43, member 3 |
1445 |
0.3 |
| chr5_119628854_119629005 | 0.34 |
Tbx3os1 |
T-box 3, opposite strand 1 |
1336 |
0.34 |
| chr9_21029266_21030734 | 0.34 |
Icam4 |
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
546 |
0.46 |
| chr13_28757451_28758760 | 0.34 |
Mir6368 |
microRNA 6368 |
47232 |
0.14 |
| chr18_32552663_32553082 | 0.33 |
Gypc |
glycophorin C |
7108 |
0.21 |
| chr2_33438550_33438701 | 0.33 |
Zbtb34 |
zinc finger and BTB domain containing 34 |
7301 |
0.15 |
| chr15_79224523_79224986 | 0.33 |
Pick1 |
protein interacting with C kinase 1 |
4419 |
0.11 |
| chr2_69341138_69341289 | 0.33 |
Abcb11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
1387 |
0.41 |
| chr7_4657800_4658971 | 0.33 |
Gm44878 |
predicted gene 44878 |
275 |
0.65 |
| chr12_110973871_110974372 | 0.33 |
Ankrd9 |
ankyrin repeat domain 9 |
4134 |
0.14 |
| chr4_126261968_126262137 | 0.32 |
Trappc3 |
trafficking protein particle complex 3 |
273 |
0.86 |
| chr16_18342108_18342456 | 0.32 |
Tango2 |
transport and golgi organization 2 |
1615 |
0.2 |
| chr10_25539576_25539847 | 0.32 |
Gm29571 |
predicted gene 29571 |
3325 |
0.21 |
| chr17_35514558_35514964 | 0.31 |
CR974473.1 |
transcription factor 19 (Tcf19) pseudogene |
1096 |
0.22 |
| chr2_31060865_31061356 | 0.31 |
Fnbp1 |
formin binding protein 1 |
5666 |
0.19 |
| chr17_84156320_84156518 | 0.31 |
Gm19696 |
predicted gene, 19696 |
7 |
0.97 |
| chr8_122658492_122658718 | 0.30 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
12139 |
0.09 |
| chr6_5803273_5803424 | 0.30 |
Dync1i1 |
dynein cytoplasmic 1 intermediate chain 1 |
45929 |
0.19 |
| chr4_119115471_119116280 | 0.30 |
Slc2a1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
6961 |
0.1 |
| chr4_6196428_6196604 | 0.30 |
Ubxn2b |
UBX domain protein 2B |
5418 |
0.26 |
| chr4_133338534_133339530 | 0.30 |
Wdtc1 |
WD and tetratricopeptide repeats 1 |
239 |
0.9 |
| chr9_66915026_66915697 | 0.29 |
Rab8b |
RAB8B, member RAS oncogene family |
4326 |
0.19 |
| chr18_62174392_62175675 | 0.29 |
Adrb2 |
adrenergic receptor, beta 2 |
4926 |
0.21 |
| chr1_161750875_161751601 | 0.29 |
Gm5049 |
predicted gene 5049 |
5182 |
0.16 |
| chr7_81652293_81652444 | 0.29 |
Gm26149 |
predicted gene, 26149 |
17360 |
0.12 |
| chr5_107224079_107224230 | 0.29 |
Gm8145 |
predicted gene 8145 |
17469 |
0.14 |
| chr11_20022067_20022218 | 0.29 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
67572 |
0.11 |
| chr15_103250315_103251530 | 0.29 |
Nfe2 |
nuclear factor, erythroid derived 2 |
543 |
0.62 |
| chr2_74663085_74663236 | 0.28 |
Evx2 |
even-skipped homeobox 2 |
3603 |
0.08 |
| chr1_132389425_132389838 | 0.27 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
687 |
0.59 |
| chr8_120454389_120454680 | 0.27 |
Gm22715 |
predicted gene, 22715 |
10985 |
0.17 |
| chr4_99030875_99031254 | 0.27 |
Angptl3 |
angiopoietin-like 3 |
110 |
0.97 |
| chr10_128821963_128822829 | 0.27 |
Sarnp |
SAP domain containing ribonucleoprotein |
464 |
0.51 |
| chr7_25088640_25088822 | 0.27 |
Mir6537 |
microRNA 6537 |
8444 |
0.11 |
| chr17_45886050_45887093 | 0.26 |
Gm41584 |
predicted gene, 41584 |
1225 |
0.42 |
| chr10_80158079_80159158 | 0.26 |
Cirbp |
cold inducible RNA binding protein |
7367 |
0.08 |
| chr1_88094712_88095046 | 0.26 |
Ugt1a7c |
UDP glucuronosyltransferase 1 family, polypeptide A7C |
183 |
0.84 |
| chr11_57649559_57650027 | 0.26 |
4933424L21Rik |
RIKEN cDNA 4933424L21 gene |
299 |
0.87 |
| chr17_79918311_79919151 | 0.25 |
Gm6552 |
predicted gene 6552 |
15841 |
0.16 |
| chr11_113779500_113779753 | 0.25 |
Gm11736 |
predicted gene 11736 |
22924 |
0.14 |
| chr4_11128483_11128667 | 0.25 |
Gm11827 |
predicted gene 11827 |
4617 |
0.13 |
| chr1_88095102_88096019 | 0.25 |
Ugt1a7c |
UDP glucuronosyltransferase 1 family, polypeptide A7C |
446 |
0.58 |
| chr17_24478260_24478723 | 0.25 |
Mlst8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
13 |
0.93 |
| chr4_136776404_136777015 | 0.25 |
Ephb2 |
Eph receptor B2 |
59134 |
0.1 |
| chr15_97785974_97786429 | 0.24 |
Slc48a1 |
solute carrier family 48 (heme transporter), member 1 |
1712 |
0.26 |
| chr2_74734325_74737080 | 0.24 |
Hoxd3 |
homeobox D3 |
813 |
0.31 |
| chr7_28307770_28307921 | 0.23 |
Timm50 |
translocase of inner mitochondrial membrane 50 |
173 |
0.87 |
| chr2_91027643_91027794 | 0.23 |
Gm13777 |
predicted gene 13777 |
3404 |
0.13 |
| chr16_7006273_7006424 | 0.22 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
63498 |
0.16 |
| chr2_38925225_38926405 | 0.22 |
Nr6a1 |
nuclear receptor subfamily 6, group A, member 1 |
402 |
0.74 |
| chr17_83684633_83684800 | 0.22 |
Mta3 |
metastasis associated 3 |
21447 |
0.21 |
| chr6_88123695_88124100 | 0.22 |
Mir6376 |
microRNA 6376 |
19639 |
0.12 |
| chr18_37955063_37956021 | 0.22 |
Rell2 |
RELT-like 2 |
17 |
0.88 |
| chr13_115376228_115376379 | 0.22 |
Gm47891 |
predicted gene, 47891 |
94102 |
0.08 |
| chr8_57611661_57611812 | 0.22 |
Gm45534 |
predicted gene 45534 |
4580 |
0.12 |
| chr13_104610150_104610741 | 0.22 |
2610204G07Rik |
RIKEN cDNA 2610204G07 gene |
74486 |
0.12 |
| chr11_121548233_121548639 | 0.22 |
Tbcd |
tubulin-specific chaperone d |
1675 |
0.38 |
| chr4_130112205_130112356 | 0.21 |
Pef1 |
penta-EF hand domain containing 1 |
4724 |
0.16 |
| chr3_96255177_96255418 | 0.21 |
Gm20627 |
predicted gene 20627 |
172 |
0.77 |
| chr6_52196485_52198020 | 0.21 |
Hoxaas3 |
Hoxa cluster antisense RNA 3 |
3872 |
0.06 |
| chr5_125465151_125466227 | 0.21 |
Gm43756 |
predicted gene 43756 |
5808 |
0.12 |
| chr10_40132385_40132661 | 0.21 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
9731 |
0.14 |
| chr15_8967409_8968940 | 0.21 |
Ranbp3l |
RAN binding protein 3-like |
172 |
0.96 |
| chr4_150717290_150717441 | 0.21 |
Gm16079 |
predicted gene 16079 |
38573 |
0.15 |
| chr5_30920335_30921507 | 0.20 |
Khk |
ketohexokinase |
510 |
0.51 |
| chr17_29005330_29006086 | 0.20 |
Stk38 |
serine/threonine kinase 38 |
2018 |
0.16 |
| chr3_126365915_126366272 | 0.20 |
Arsj |
arylsulfatase J |
2409 |
0.33 |
| chr6_88925895_88926135 | 0.20 |
Gm44178 |
predicted gene, 44178 |
6307 |
0.13 |
| chr4_141159081_141159521 | 0.20 |
Fbxo42 |
F-box protein 42 |
11379 |
0.12 |
| chr7_45093013_45094086 | 0.20 |
Rcn3 |
reticulocalbin 3, EF-hand calcium binding domain |
1328 |
0.13 |
| chr2_90942971_90943203 | 0.20 |
Celf1 |
CUGBP, Elav-like family member 1 |
2544 |
0.17 |
| chr18_21152486_21153141 | 0.20 |
Gm6378 |
predicted pseudogene 6378 |
75704 |
0.09 |
| chr3_95141713_95142452 | 0.20 |
Tnfaip8l2 |
tumor necrosis factor, alpha-induced protein 8-like 2 |
278 |
0.74 |
| chr16_86516621_86516772 | 0.20 |
Gm49572 |
predicted gene, 49572 |
242203 |
0.02 |
| chr11_11831905_11832402 | 0.20 |
Ddc |
dopa decarboxylase |
4127 |
0.22 |
| chr2_132518446_132518597 | 0.19 |
Gm14095 |
predicted gene 14095 |
17373 |
0.15 |
| chr17_29489359_29490304 | 0.19 |
Pim1 |
proviral integration site 1 |
922 |
0.36 |
| chr3_102152409_102152561 | 0.19 |
Casq2 |
calsequestrin 2 |
8434 |
0.13 |
| chr7_80628877_80629821 | 0.19 |
Crtc3 |
CREB regulated transcription coactivator 3 |
277 |
0.89 |
| chr8_12548291_12548587 | 0.19 |
Spaca7 |
sperm acrosome associated 7 |
24590 |
0.14 |
| chr14_27338331_27338622 | 0.19 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
2410 |
0.32 |
| chr4_141286800_141287991 | 0.19 |
Gm13056 |
predicted gene 13056 |
8956 |
0.11 |
| chr7_27957807_27957958 | 0.19 |
Gm10651 |
predicted pseudogene 10651 |
8119 |
0.11 |
| chr6_48504124_48504282 | 0.19 |
Zfp862-ps |
zinc finger protein 862, pseudogene |
134 |
0.91 |
| chr19_6280905_6281787 | 0.19 |
Ehd1 |
EH-domain containing 1 |
4421 |
0.08 |
| chr11_55458328_55458479 | 0.19 |
Atox1 |
antioxidant 1 copper chaperone |
2794 |
0.16 |
| chr16_93744711_93744862 | 0.18 |
Dop1b |
DOP1 leucine zipper like protein B |
14687 |
0.16 |
| chr11_95145006_95146328 | 0.18 |
Dlx4 |
distal-less homeobox 4 |
134 |
0.86 |
| chr7_141276058_141276884 | 0.18 |
Cdhr5 |
cadherin-related family member 5 |
303 |
0.75 |
| chr11_55077185_55077593 | 0.18 |
Ccdc69 |
coiled-coil domain containing 69 |
718 |
0.58 |
| chr11_61686935_61687279 | 0.18 |
Fam83g |
family with sequence similarity 83, member G |
2688 |
0.23 |
| chr18_11049995_11051717 | 0.18 |
Gata6os |
GATA binding protein 6, opposite strand |
631 |
0.64 |
| chr5_123147784_123150032 | 0.17 |
Setd1b |
SET domain containing 1B |
5951 |
0.08 |
| chr4_133041805_133043935 | 0.17 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
3377 |
0.24 |
| chr13_93428749_93428900 | 0.17 |
Gm18960 |
predicted gene, 18960 |
7508 |
0.15 |
| chr10_91542314_91542465 | 0.17 |
Gm47084 |
predicted gene, 47084 |
94009 |
0.08 |
| chr5_125390257_125390622 | 0.17 |
Ubc |
ubiquitin C |
237 |
0.84 |
| chr2_170201888_170202331 | 0.17 |
Zfp217 |
zinc finger protein 217 |
54006 |
0.14 |
| chr3_34834735_34834886 | 0.17 |
Gm21388 |
predicted gene, 21388 |
2340 |
0.34 |
| chr17_40878627_40878778 | 0.17 |
9130008F23Rik |
RIKEN cDNA 9130008F23 gene |
1856 |
0.26 |
| chr2_26138540_26139205 | 0.16 |
Tmem250-ps |
transmembrane protein 250, pseudogene |
70 |
0.8 |
| chr11_96311089_96312545 | 0.16 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
4907 |
0.07 |
| chr11_96790204_96790385 | 0.16 |
Cbx1 |
chromobox 1 |
1045 |
0.36 |
| chr10_45467792_45468248 | 0.16 |
Lin28b |
lin-28 homolog B (C. elegans) |
2181 |
0.36 |
| chr5_112344442_112344593 | 0.16 |
Hps4 |
HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 |
1417 |
0.21 |
| chr3_84445916_84446485 | 0.16 |
Fhdc1 |
FH2 domain containing 1 |
7563 |
0.24 |
| chr7_110921038_110921189 | 0.16 |
Mrvi1 |
MRV integration site 1 |
2590 |
0.25 |
| chr6_49210050_49211632 | 0.16 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3320 |
0.2 |
| chr19_14426285_14426840 | 0.16 |
Tle4 |
transducin-like enhancer of split 4 |
168977 |
0.03 |
| chr18_61954214_61954483 | 0.16 |
Sh3tc2 |
SH3 domain and tetratricopeptide repeats 2 |
1264 |
0.47 |
| chr13_107074847_107075087 | 0.15 |
Gm31452 |
predicted gene, 31452 |
11272 |
0.17 |
| chr3_65658538_65659742 | 0.15 |
Mir8120 |
microRNA 8120 |
148 |
0.94 |
| chr11_69419130_69420636 | 0.15 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
6208 |
0.09 |
| chr9_71213599_71213944 | 0.15 |
Aldh1a2 |
aldehyde dehydrogenase family 1, subfamily A2 |
2018 |
0.34 |
| chr11_95826836_95827629 | 0.15 |
Phospho1 |
phosphatase, orphan 1 |
2732 |
0.15 |
| chr9_65193224_65193667 | 0.15 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
1908 |
0.22 |
| chr6_84882813_84883593 | 0.15 |
Exoc6b |
exocyst complex component 6B |
10785 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.4 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.3 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 | 0.5 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 1.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 1.8 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.3 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.3 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.7 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) |
| 0.0 | 0.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |