Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
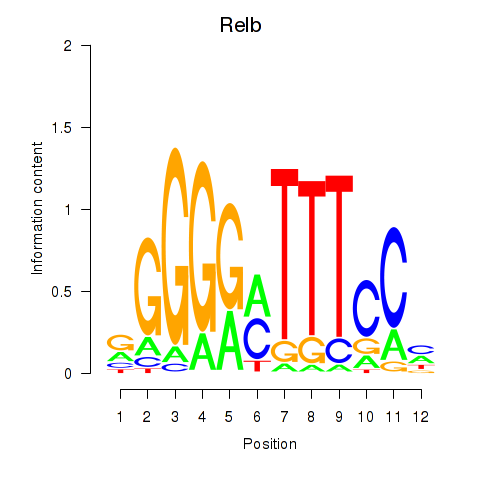
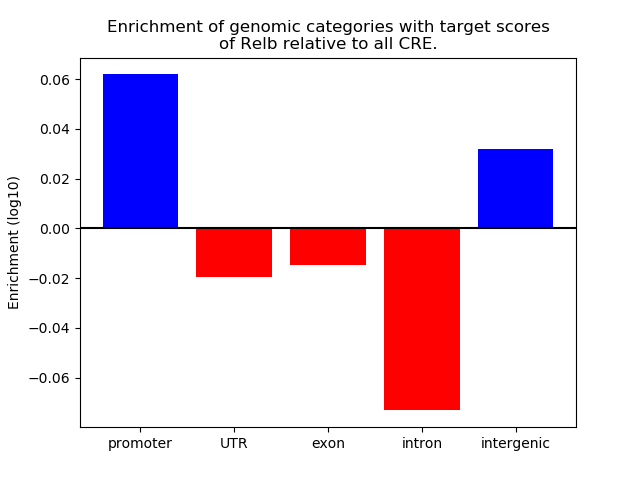
Results for Relb
Z-value: 0.51
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.10 | avian reticuloendotheliosis viral (v-rel) oncogene related B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_19628251_19629412 | Relb | 47 | 0.942289 | -0.49 | 7.6e-05 | Click! |
| chr7_19629476_19629768 | Relb | 184 | 0.877869 | -0.46 | 1.9e-04 | Click! |
| chr7_19631359_19631510 | Relb | 1996 | 0.157446 | 0.13 | 3.4e-01 | Click! |
| chr7_19622440_19622591 | Relb | 5919 | 0.091779 | 0.10 | 4.6e-01 | Click! |
| chr7_19632220_19632371 | Relb | 2857 | 0.118901 | -0.08 | 5.2e-01 | Click! |
Activity of the Relb motif across conditions
Conditions sorted by the z-value of the Relb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
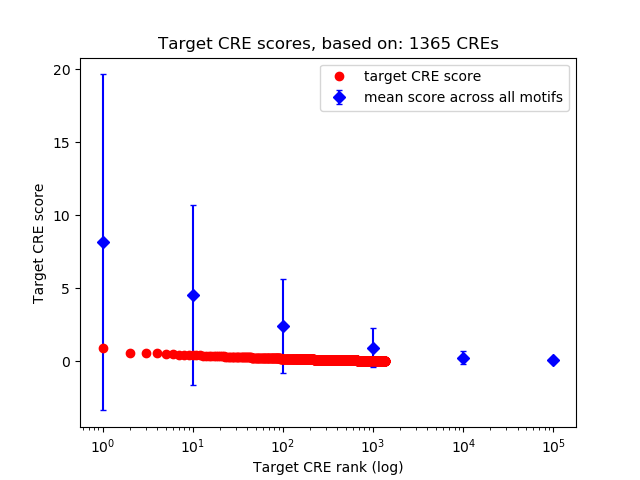
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_57320946_57324000 | 0.89 |
Hand2os1 |
Hand2, opposite strand 1 |
1245 |
0.3 |
| chr19_31624383_31624534 | 0.60 |
Prkg1 |
protein kinase, cGMP-dependent, type I |
39912 |
0.22 |
| chr7_132270093_132270244 | 0.56 |
Chst15 |
carbohydrate sulfotransferase 15 |
8457 |
0.18 |
| chr10_79664388_79664620 | 0.54 |
Madcam1 |
mucosal vascular addressin cell adhesion molecule 1 |
55 |
0.93 |
| chr5_106457979_106459205 | 0.52 |
Barhl2 |
BarH like homeobox 2 |
426 |
0.82 |
| chr12_27333974_27334218 | 0.51 |
Sox11 |
SRY (sex determining region Y)-box 11 |
8478 |
0.29 |
| chr2_165741610_165742546 | 0.46 |
Eya2 |
EYA transcriptional coactivator and phosphatase 2 |
19746 |
0.2 |
| chr4_150651111_150652374 | 0.44 |
Slc45a1 |
solute carrier family 45, member 1 |
355 |
0.88 |
| chr11_36438334_36438485 | 0.44 |
Tenm2 |
teneurin transmembrane protein 2 |
6133 |
0.33 |
| chr15_102959654_102960750 | 0.41 |
Hoxc11 |
homeobox C11 |
5775 |
0.08 |
| chr11_44707619_44708111 | 0.40 |
Gm12158 |
predicted gene 12158 |
45189 |
0.16 |
| chr14_122480308_122481080 | 0.40 |
Zic2 |
zinc finger protein of the cerebellum 2 |
2594 |
0.16 |
| chr11_96272013_96272961 | 0.39 |
Hoxb9 |
homeobox B9 |
964 |
0.29 |
| chr10_42581935_42584872 | 0.39 |
Nr2e1 |
nuclear receptor subfamily 2, group E, member 1 |
229 |
0.69 |
| chr3_116157583_116157734 | 0.38 |
Gm26544 |
predicted gene, 26544 |
7179 |
0.14 |
| chr12_55321703_55321892 | 0.37 |
Prorp |
protein only RNase P catalytic subunit |
18511 |
0.13 |
| chr7_80058773_80058974 | 0.36 |
Gm21057 |
predicted gene, 21057 |
16374 |
0.1 |
| chr6_30544847_30545729 | 0.36 |
Cpa2 |
carboxypeptidase A2, pancreatic |
803 |
0.52 |
| chr4_106976768_106976971 | 0.33 |
Ssbp3 |
single-stranded DNA binding protein 3 |
10443 |
0.19 |
| chr1_133050009_133050364 | 0.33 |
Pik3c2b |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
4174 |
0.17 |
| chr8_61360117_61360268 | 0.33 |
Sh3rf1 |
SH3 domain containing ring finger 1 |
1397 |
0.39 |
| chr19_43606004_43606218 | 0.33 |
Gm20467 |
predicted gene 20467 |
4514 |
0.13 |
| chr4_139578966_139579453 | 0.32 |
Iffo2 |
intermediate filament family orphan 2 |
4489 |
0.17 |
| chr2_105125289_105128976 | 0.32 |
Wt1 |
Wilms tumor 1 homolog |
78 |
0.91 |
| chr14_79290059_79290488 | 0.32 |
Rgcc |
regulator of cell cycle |
11372 |
0.16 |
| chr15_98021856_98022007 | 0.32 |
Col2a1 |
collagen, type II, alpha 1 |
17236 |
0.13 |
| chr10_22818712_22820254 | 0.31 |
Tcf21 |
transcription factor 21 |
676 |
0.65 |
| chr11_70043720_70044094 | 0.31 |
Dlg4 |
discs large MAGUK scaffold protein 4 |
290 |
0.77 |
| chr12_111166036_111167450 | 0.31 |
Traf3 |
TNF receptor-associated factor 3 |
195 |
0.94 |
| chr5_123181886_123182179 | 0.31 |
Hpd |
4-hydroxyphenylpyruvic acid dioxygenase |
35 |
0.95 |
| chr6_52202371_52204739 | 0.31 |
Hoxa5 |
homeobox A5 |
1032 |
0.2 |
| chr2_146834060_146838027 | 0.31 |
Gm14114 |
predicted gene 14114 |
3689 |
0.27 |
| chr13_73268193_73269257 | 0.30 |
Gm48000 |
predicted gene, 48000 |
4505 |
0.18 |
| chr16_97607830_97607981 | 0.28 |
Tmprss2 |
transmembrane protease, serine 2 |
3064 |
0.28 |
| chr7_80404041_80404273 | 0.28 |
Gm44851 |
predicted gene 44851 |
112 |
0.9 |
| chr3_78901151_78901971 | 0.28 |
Gm5277 |
predicted gene 5277 |
9139 |
0.18 |
| chr5_135040050_135040229 | 0.28 |
Stx1a |
syntaxin 1A (brain) |
921 |
0.33 |
| chr16_38360437_38360588 | 0.27 |
Popdc2 |
popeye domain containing 2 |
1697 |
0.25 |
| chr19_44755099_44757716 | 0.27 |
Pax2 |
paired box 2 |
362 |
0.83 |
| chr2_70558567_70559918 | 0.27 |
Gad1 |
glutamate decarboxylase 1 |
2800 |
0.19 |
| chr6_99200508_99201678 | 0.27 |
Foxp1 |
forkhead box P1 |
38075 |
0.21 |
| chr7_74006482_74006835 | 0.26 |
St8sia2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
7016 |
0.2 |
| chr2_180502555_180502814 | 0.26 |
Ntsr1 |
neurotensin receptor 1 |
2457 |
0.25 |
| chr5_75084798_75085749 | 0.26 |
Gsx2 |
GS homeobox 2 |
9671 |
0.12 |
| chr9_50850324_50851021 | 0.26 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
5371 |
0.16 |
| chr9_5308602_5309732 | 0.26 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
302 |
0.91 |
| chr18_75405225_75405894 | 0.26 |
Smad7 |
SMAD family member 7 |
30645 |
0.18 |
| chr13_71104368_71105400 | 0.25 |
Mir466f-4 |
microRNA 466f-4 |
2205 |
0.39 |
| chr14_60735927_60737075 | 0.25 |
Spata13 |
spermatogenesis associated 13 |
3595 |
0.24 |
| chr4_135956691_135956842 | 0.25 |
Hmgcl |
3-hydroxy-3-methylglutaryl-Coenzyme A lyase |
2961 |
0.12 |
| chr7_35186002_35186989 | 0.25 |
Slc7a10 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 |
13 |
0.96 |
| chr6_129449977_129450616 | 0.25 |
Clec1a |
C-type lectin domain family 1, member a |
1539 |
0.24 |
| chr13_83732205_83734272 | 0.24 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
672 |
0.58 |
| chr9_119979553_119979704 | 0.24 |
Csrnp1 |
cysteine-serine-rich nuclear protein 1 |
2293 |
0.16 |
| chr7_138199866_138200038 | 0.24 |
Tcerg1l |
transcription elongation regulator 1-like |
71377 |
0.12 |
| chr19_59467055_59468700 | 0.23 |
Emx2 |
empty spiracles homeobox 2 |
5075 |
0.18 |
| chr3_20147878_20148137 | 0.23 |
Gyg |
glycogenin |
7062 |
0.21 |
| chr17_26838158_26839992 | 0.22 |
Nkx2-5 |
NK2 homeobox 5 |
2490 |
0.17 |
| chr11_103770931_103772004 | 0.22 |
Wnt3 |
wingless-type MMTV integration site family, member 3 |
2683 |
0.23 |
| chr4_138469035_138469593 | 0.22 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
15000 |
0.15 |
| chr9_111021840_111021991 | 0.22 |
Ltf |
lactotransferrin |
501 |
0.64 |
| chr19_55899988_55900963 | 0.22 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
2166 |
0.4 |
| chr9_64240654_64240822 | 0.22 |
Uchl4 |
ubiquitin carboxyl-terminal esterase L4 |
5537 |
0.1 |
| chr7_142662290_142664788 | 0.22 |
Igf2os |
insulin-like growth factor 2, opposite strand |
1599 |
0.21 |
| chr11_32221179_32222558 | 0.22 |
Rhbdf1 |
rhomboid 5 homolog 1 |
376 |
0.76 |
| chr11_96306504_96308444 | 0.22 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
564 |
0.48 |
| chr8_124249985_124250235 | 0.22 |
Galnt2 |
polypeptide N-acetylgalactosaminyltransferase 2 |
18602 |
0.17 |
| chr6_52244514_52245126 | 0.22 |
Hoxa11os |
homeobox A11, opposite strand |
423 |
0.51 |
| chr19_41699162_41699313 | 0.21 |
Slit1 |
slit guidance ligand 1 |
44249 |
0.14 |
| chr11_114652641_114652923 | 0.21 |
Gm11689 |
predicted gene 11689 |
11826 |
0.16 |
| chr19_44758783_44762005 | 0.21 |
Pax2 |
paired box 2 |
479 |
0.75 |
| chr5_34183436_34183719 | 0.21 |
Mxd4 |
Max dimerization protein 4 |
793 |
0.44 |
| chr6_125363043_125363874 | 0.21 |
Tnfrsf1a |
tumor necrosis factor receptor superfamily, member 1a |
3031 |
0.17 |
| chr2_30872666_30872817 | 0.21 |
Prrx2 |
paired related homeobox 2 |
6578 |
0.14 |
| chr9_79919650_79920321 | 0.21 |
Gm3211 |
predicted gene 3211 |
6785 |
0.21 |
| chr3_68470131_68470282 | 0.20 |
Schip1 |
schwannomin interacting protein 1 |
2023 |
0.4 |
| chr11_67921338_67922636 | 0.20 |
Usp43 |
ubiquitin specific peptidase 43 |
166 |
0.74 |
| chr7_18949615_18951696 | 0.20 |
Nova2 |
NOVA alternative splicing regulator 2 |
24767 |
0.07 |
| chr6_52245764_52247193 | 0.20 |
Hoxa11os |
homeobox A11, opposite strand |
257 |
0.65 |
| chr9_115391502_115391653 | 0.20 |
Gm25510 |
predicted gene, 25510 |
9478 |
0.12 |
| chr2_70127042_70128123 | 0.20 |
Myo3b |
myosin IIIB |
31284 |
0.2 |
| chr18_37742006_37744626 | 0.20 |
Pcdhgb6 |
protocadherin gamma subfamily B, 6 |
1222 |
0.15 |
| chr9_63144078_63146888 | 0.20 |
Skor1 |
SKI family transcriptional corepressor 1 |
1497 |
0.38 |
| chr5_75148315_75152589 | 0.20 |
Pdgfra |
platelet derived growth factor receptor, alpha polypeptide |
1840 |
0.2 |
| chr6_48025548_48025741 | 0.20 |
Zfp777 |
zinc finger protein 777 |
18599 |
0.12 |
| chr3_126996871_126998855 | 0.20 |
Ank2 |
ankyrin 2, brain |
575 |
0.63 |
| chr15_84301565_84301928 | 0.19 |
Gm22890 |
predicted gene, 22890 |
21606 |
0.13 |
| chr9_44733806_44735185 | 0.19 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
703 |
0.43 |
| chr2_31547915_31548066 | 0.19 |
Gm13426 |
predicted gene 13426 |
4705 |
0.17 |
| chr17_65905371_65905522 | 0.19 |
Gm35550 |
predicted gene, 35550 |
86 |
0.95 |
| chr11_118998485_119000015 | 0.19 |
Enpp7 |
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
11034 |
0.16 |
| chr4_32923224_32923939 | 0.19 |
Ankrd6 |
ankyrin repeat domain 6 |
76 |
0.97 |
| chr3_38568695_38568846 | 0.19 |
Gm7824 |
predicted gene 7824 |
10102 |
0.18 |
| chr15_103011882_103012844 | 0.19 |
Hoxc5 |
homeobox C5 |
1452 |
0.19 |
| chr9_15534305_15534456 | 0.19 |
Smco4 |
single-pass membrane protein with coiled-coil domains 4 |
13521 |
0.17 |
| chr12_26534879_26535030 | 0.19 |
Gm46344 |
predicted gene, 46344 |
48163 |
0.11 |
| chr9_31120528_31120906 | 0.19 |
St14 |
suppression of tumorigenicity 14 (colon carcinoma) |
11010 |
0.15 |
| chr4_149571821_149571972 | 0.19 |
Gm13066 |
predicted gene 13066 |
4106 |
0.14 |
| chr11_117866335_117866496 | 0.19 |
Gm11725 |
predicted gene 11725 |
4835 |
0.1 |
| chr19_5930278_5930429 | 0.19 |
Cdc42ep2 |
CDC42 effector protein (Rho GTPase binding) 2 |
5537 |
0.08 |
| chr1_130550439_130551075 | 0.19 |
Gm23623 |
predicted gene, 23623 |
44919 |
0.1 |
| chr8_121398391_121398555 | 0.19 |
Gm26747 |
predicted gene, 26747 |
22765 |
0.16 |
| chr18_37720288_37722950 | 0.18 |
Pcdhgb4 |
protocadherin gamma subfamily B, 4 |
1236 |
0.14 |
| chr11_45868095_45868246 | 0.18 |
Clint1 |
clathrin interactor 1 |
14227 |
0.15 |
| chr13_30748912_30751060 | 0.18 |
Irf4 |
interferon regulatory factor 4 |
679 |
0.71 |
| chr18_35627084_35627237 | 0.18 |
Slc23a1 |
solute carrier family 23 (nucleobase transporters), member 1 |
36 |
0.94 |
| chr4_125122525_125123430 | 0.18 |
Zc3h12a |
zinc finger CCCH type containing 12A |
457 |
0.77 |
| chr16_43303539_43303924 | 0.18 |
Gm37946 |
predicted gene, 37946 |
4550 |
0.21 |
| chr6_134888545_134888805 | 0.18 |
Gpr19 |
G protein-coupled receptor 19 |
843 |
0.44 |
| chr15_83677852_83678399 | 0.18 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
17645 |
0.2 |
| chr14_101886945_101887764 | 0.18 |
Lmo7 |
LIM domain only 7 |
3235 |
0.35 |
| chr12_29937955_29939229 | 0.18 |
Pxdn |
peroxidasin |
542 |
0.84 |
| chr4_136031178_136031911 | 0.18 |
Rpl11 |
ribosomal protein L11 |
2746 |
0.14 |
| chr13_83737592_83739114 | 0.18 |
Gm33366 |
predicted gene, 33366 |
182 |
0.66 |
| chr4_155451781_155451950 | 0.17 |
Cfap74 |
cilia and flagella associated protein 74 |
295 |
0.84 |
| chr16_22161383_22162159 | 0.17 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
321 |
0.89 |
| chr11_116194935_116195163 | 0.17 |
Acox1 |
acyl-Coenzyme A oxidase 1, palmitoyl |
3680 |
0.11 |
| chr4_44694373_44694748 | 0.17 |
5730488B01Rik |
RIKEN cDNA 5730488B01 gene |
8240 |
0.14 |
| chr11_101418651_101418802 | 0.17 |
Mir6929 |
microRNA 6929 |
527 |
0.44 |
| chr10_79965080_79965231 | 0.17 |
Wdr18 |
WD repeat domain 18 |
349 |
0.68 |
| chr6_52167312_52169129 | 0.17 |
Hoxaas2 |
Hoxa cluster antisense RNA 2 |
290 |
0.7 |
| chr15_102950417_102951549 | 0.17 |
Hotair |
HOX transcript antisense RNA (non-protein coding) |
3253 |
0.11 |
| chr10_50895645_50896998 | 0.17 |
Sim1 |
single-minded family bHLH transcription factor 1 |
670 |
0.79 |
| chr3_148988258_148989425 | 0.17 |
Gm43573 |
predicted gene 43573 |
311 |
0.63 |
| chr7_70359645_70360747 | 0.17 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
275 |
0.86 |
| chr8_120833068_120833219 | 0.17 |
Gm26878 |
predicted gene, 26878 |
47063 |
0.12 |
| chr11_96279294_96280222 | 0.16 |
Hoxb8 |
homeobox B8 |
2147 |
0.12 |
| chr7_116034244_116034507 | 0.16 |
Sox6 |
SRY (sex determining region Y)-box 6 |
138 |
0.94 |
| chr1_133068155_133068426 | 0.16 |
Gm28609 |
predicted gene 28609 |
115 |
0.72 |
| chr1_75345328_75345561 | 0.16 |
Des |
desmin |
14885 |
0.09 |
| chr19_53710843_53711000 | 0.16 |
Rbm20 |
RNA binding motif protein 20 |
33615 |
0.15 |
| chr4_154948550_154949380 | 0.16 |
Hes5 |
hes family bHLH transcription factor 5 |
11958 |
0.11 |
| chr2_130638035_130638438 | 0.16 |
Lzts3 |
leucine zipper, putative tumor suppressor family member 3 |
141 |
0.92 |
| chr15_103002581_103003309 | 0.16 |
Hoxc6 |
homeobox C6 |
2416 |
0.13 |
| chr2_30074557_30074708 | 0.16 |
Pkn3 |
protein kinase N3 |
3052 |
0.14 |
| chr6_51320497_51320934 | 0.16 |
Gm32479 |
predicted gene, 32479 |
32860 |
0.15 |
| chr5_113504103_113504254 | 0.16 |
Wscd2 |
WSC domain containing 2 |
13426 |
0.22 |
| chr10_4080039_4080600 | 0.15 |
Gm25515 |
predicted gene, 25515 |
23994 |
0.18 |
| chr17_21753488_21754324 | 0.15 |
Zfp229 |
zinc finger protein 229 |
17994 |
0.11 |
| chr10_10546593_10546906 | 0.15 |
Rab32 |
RAB32, member RAS oncogene family |
11432 |
0.2 |
| chr9_34871552_34871717 | 0.15 |
Kirrel3os |
kirre like nephrin family adhesion molecule 3, opposite strand |
57875 |
0.13 |
| chr7_70287627_70288555 | 0.15 |
Gm29327 |
predicted gene 29327 |
38315 |
0.11 |
| chr2_174049677_174049894 | 0.15 |
Vamp7-ps |
vesicle-associated membrane protein 7, pseudogene |
2512 |
0.27 |
| chr15_67040638_67040789 | 0.15 |
Gm31342 |
predicted gene, 31342 |
655 |
0.76 |
| chr4_32923966_32924256 | 0.15 |
Ankrd6 |
ankyrin repeat domain 6 |
606 |
0.7 |
| chr2_113757445_113759087 | 0.15 |
Grem1 |
gremlin 1, DAN family BMP antagonist |
380 |
0.87 |
| chr13_56250027_56250511 | 0.15 |
Neurog1 |
neurogenin 1 |
1894 |
0.26 |
| chr17_79069824_79069975 | 0.15 |
Qpct |
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
7103 |
0.18 |
| chr18_37763101_37764236 | 0.15 |
Pcdhgb8 |
protocadherin gamma subfamily B, 8 |
1867 |
0.09 |
| chr11_101332029_101333108 | 0.15 |
Aoc3 |
amine oxidase, copper containing 3 |
1499 |
0.15 |
| chr17_29447192_29447381 | 0.15 |
Gm36199 |
predicted gene, 36199 |
1381 |
0.3 |
| chr2_132305611_132305762 | 0.15 |
Cds2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
4390 |
0.21 |
| chr9_108296727_108297858 | 0.15 |
Amt |
aminomethyltransferase |
370 |
0.7 |
| chr1_9788025_9788203 | 0.15 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
4336 |
0.15 |
| chr10_18470218_18471289 | 0.14 |
Nhsl1 |
NHS-like 1 |
772 |
0.72 |
| chr11_95594860_95595011 | 0.14 |
Ngfr |
nerve growth factor receptor (TNFR superfamily, member 16) |
7200 |
0.17 |
| chr13_47645272_47645423 | 0.14 |
4930471G24Rik |
RIKEN cDNA 4930471G24 gene |
22203 |
0.27 |
| chr18_60649080_60649231 | 0.14 |
Synpo |
synaptopodin |
836 |
0.6 |
| chr2_10153329_10154684 | 0.14 |
Itih5 |
inter-alpha (globulin) inhibitor H5 |
435 |
0.76 |
| chr2_74734325_74737080 | 0.14 |
Hoxd3 |
homeobox D3 |
813 |
0.31 |
| chr2_6881874_6882908 | 0.14 |
Gm13389 |
predicted gene 13389 |
1879 |
0.3 |
| chr2_163637671_163638403 | 0.14 |
0610039K10Rik |
RIKEN cDNA 0610039K10 gene |
6813 |
0.13 |
| chr5_100358403_100358554 | 0.14 |
Gm8091 |
predicted gene 8091 |
3359 |
0.21 |
| chr17_34905022_34905208 | 0.14 |
Ehmt2 |
euchromatic histone lysine N-methyltransferase 2 |
88 |
0.89 |
| chr1_128643284_128643435 | 0.14 |
Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
51066 |
0.13 |
| chr6_52313324_52314938 | 0.14 |
Evx1 |
even-skipped homeobox 1 |
579 |
0.38 |
| chr17_57148993_57149872 | 0.14 |
Cd70 |
CD70 antigen |
345 |
0.79 |
| chr2_116071729_116072658 | 0.14 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
3903 |
0.19 |
| chr2_155380370_155380655 | 0.14 |
Trp53inp2 |
transformation related protein 53 inducible nuclear protein 2 |
547 |
0.69 |
| chr1_152086908_152087700 | 0.14 |
1700025G04Rik |
RIKEN cDNA 1700025G04 gene |
2666 |
0.35 |
| chr2_25213719_25213959 | 0.14 |
Stpg3 |
sperm tail PG rich repeat containing 3 |
188 |
0.8 |
| chr8_34325722_34326983 | 0.14 |
Gm4889 |
predicted gene 4889 |
1558 |
0.28 |
| chr14_31212442_31212593 | 0.14 |
Tnnc1 |
troponin C, cardiac/slow skeletal |
4185 |
0.12 |
| chr15_85615061_85615935 | 0.14 |
Gm49539 |
predicted gene, 49539 |
30585 |
0.12 |
| chr2_18038889_18039040 | 0.14 |
A930004D18Rik |
RIKEN cDNA A930004D18 gene |
1227 |
0.31 |
| chr6_65674892_65675628 | 0.13 |
Ndnf |
neuron-derived neurotrophic factor |
3670 |
0.28 |
| chr9_4794566_4795419 | 0.13 |
Gria4 |
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
527 |
0.88 |
| chr1_132194845_132194996 | 0.13 |
Mir135b |
microRNA 135b |
3168 |
0.14 |
| chr1_138171970_138172121 | 0.13 |
Ptprc |
protein tyrosine phosphatase, receptor type, C |
3144 |
0.18 |
| chr17_24657836_24658944 | 0.13 |
Npw |
neuropeptide W |
67 |
0.92 |
| chr18_37690944_37692448 | 0.13 |
Pcdhgb2 |
protocadherin gamma subfamily B, 2 |
1861 |
0.09 |
| chr11_101965649_101967097 | 0.13 |
Sost |
sclerostin |
642 |
0.52 |
| chr3_89965873_89966024 | 0.13 |
Gm24046 |
predicted gene, 24046 |
2215 |
0.13 |
| chr16_69721275_69721426 | 0.13 |
4930428D20Rik |
RIKEN cDNA 4930428D20 gene |
79031 |
0.12 |
| chr7_19009114_19009265 | 0.13 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
5145 |
0.08 |
| chr2_165233771_165234688 | 0.13 |
Cdh22 |
cadherin 22 |
624 |
0.69 |
| chr2_93189809_93190397 | 0.13 |
Trp53i11 |
transformation related protein 53 inducible protein 11 |
912 |
0.64 |
| chr10_127048288_127050177 | 0.13 |
Cyp27b1 |
cytochrome P450, family 27, subfamily b, polypeptide 1 |
561 |
0.49 |
| chr6_52259871_52261414 | 0.13 |
Hoxa13 |
homeobox A13 |
160 |
0.84 |
| chr3_89234782_89235064 | 0.13 |
Muc1 |
mucin 1, transmembrane |
3452 |
0.07 |
| chr1_153048305_153048494 | 0.13 |
Nmnat2 |
nicotinamide nucleotide adenylyltransferase 2 |
15025 |
0.19 |
| chr19_45017873_45019099 | 0.13 |
Lzts2 |
leucine zipper, putative tumor suppressor 2 |
274 |
0.83 |
| chr15_91571286_91571688 | 0.13 |
Slc2a13 |
solute carrier family 2 (facilitated glucose transporter), member 13 |
1707 |
0.3 |
| chr13_49700528_49700679 | 0.13 |
Iars |
isoleucine-tRNA synthetase |
5203 |
0.12 |
| chr3_98283534_98283685 | 0.13 |
Hmgcs2 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
3174 |
0.18 |
| chr8_35699415_35699737 | 0.13 |
1700015I17Rik |
RIKEN cDNA 1700015I17 gene |
20373 |
0.14 |
| chr9_70934045_70934653 | 0.13 |
Lipc |
lipase, hepatic |
264 |
0.92 |
| chr17_28510395_28511318 | 0.13 |
Fkbp5 |
FK506 binding protein 5 |
501 |
0.58 |
| chr11_119237088_119237239 | 0.13 |
Ccdc40 |
coiled-coil domain containing 40 |
1555 |
0.27 |
| chr12_115920443_115920840 | 0.13 |
Gm42990 |
predicted gene 42990 |
12850 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1900200 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.1 | 0.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.2 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0003211 | cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:2000739 | regulation of mesenchymal stem cell differentiation(GO:2000739) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.0 | GO:0072177 | mesonephric duct development(GO:0072177) |
| 0.0 | 0.1 | GO:0072497 | mesenchymal stem cell differentiation(GO:0072497) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.5 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.0 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0060897 | neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0072148 | epithelial cell fate commitment(GO:0072148) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.0 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis(GO:1902338) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.0 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.1 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |