Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
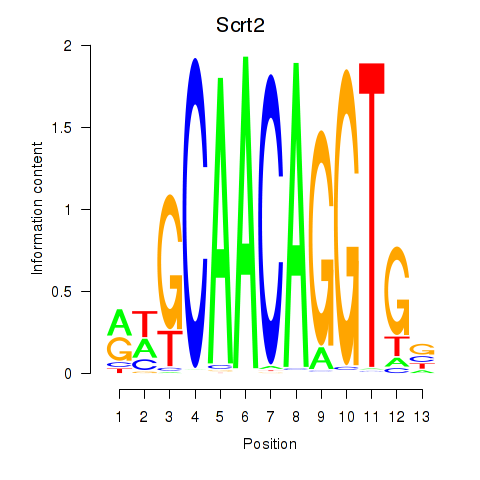
Results for Scrt2
Z-value: 0.57
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSMUSG00000060257.2 | scratch family zinc finger 2 |
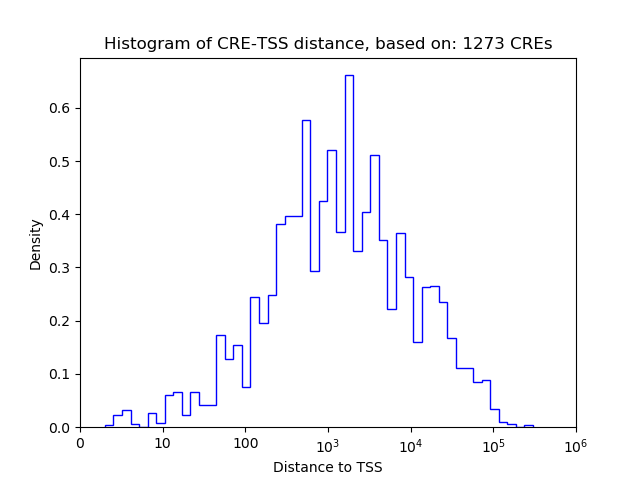
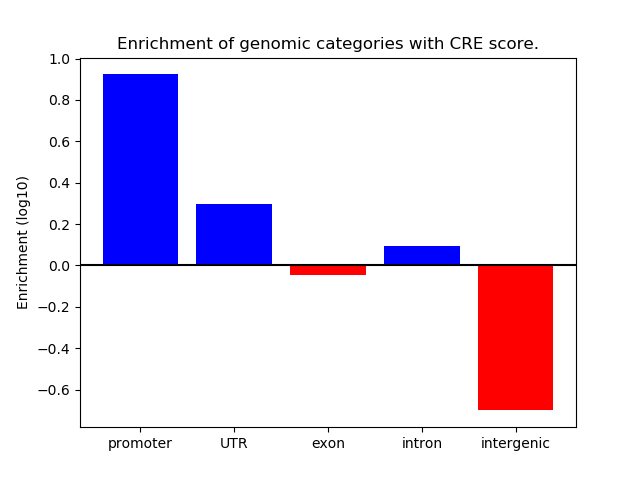
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_152084535_152084698 | Scrt2 | 3087 | 0.185283 | -0.49 | 5.9e-05 | Click! |
| chr2_152086279_152087493 | Scrt2 | 5357 | 0.151051 | -0.48 | 9.0e-05 | Click! |
| chr2_152084019_152084489 | Scrt2 | 2725 | 0.198688 | -0.45 | 3.1e-04 | Click! |
| chr2_152083384_152083946 | Scrt2 | 2136 | 0.234271 | -0.41 | 1.0e-03 | Click! |
| chr2_152085191_152085589 | Scrt2 | 3861 | 0.167396 | -0.41 | 1.2e-03 | Click! |
Activity of the Scrt2 motif across conditions
Conditions sorted by the z-value of the Scrt2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
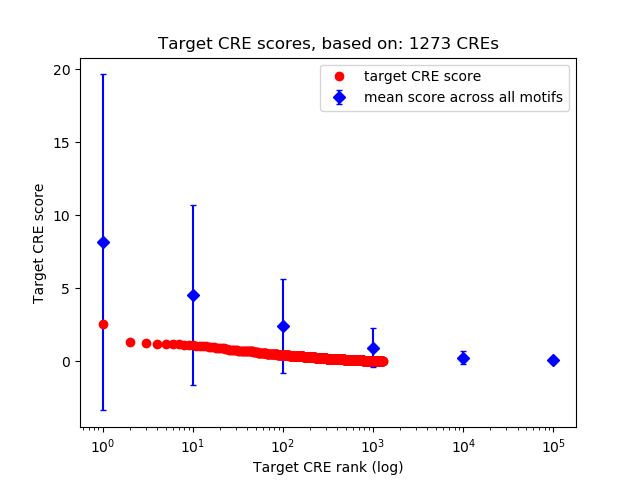
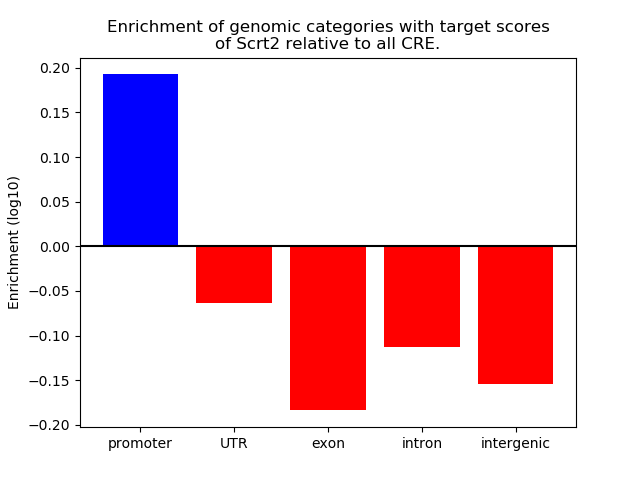
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_114051146_114052179 | 2.57 |
Actc1 |
actin, alpha, cardiac muscle 1 |
1225 |
0.4 |
| chr16_14707769_14707934 | 1.32 |
Snai2 |
snail family zinc finger 2 |
1999 |
0.4 |
| chr11_106381435_106381885 | 1.23 |
Icam2 |
intercellular adhesion molecule 2 |
859 |
0.5 |
| chr18_61652982_61653392 | 1.20 |
Mir143 |
microRNA 143 |
3929 |
0.13 |
| chr14_101884566_101885570 | 1.19 |
Lmo7 |
LIM domain only 7 |
949 |
0.69 |
| chr13_94062433_94063410 | 1.17 |
Lhfpl2 |
lipoma HMGIC fusion partner-like 2 |
5089 |
0.2 |
| chr7_46237720_46238666 | 1.16 |
Ush1c |
USH1 protein network component harmonin |
271 |
0.88 |
| chr12_59129039_59129247 | 1.14 |
Mia2 |
MIA SH3 domain ER export factor 2 |
577 |
0.66 |
| chr18_11053961_11055254 | 1.11 |
Gata6 |
GATA binding protein 6 |
431 |
0.83 |
| chr7_19399234_19400287 | 1.09 |
Klc3 |
kinesin light chain 3 |
118 |
0.89 |
| chr8_57213759_57214585 | 1.07 |
Gm17994 |
predicted gene, 17994 |
77880 |
0.08 |
| chr1_155990531_155990714 | 1.05 |
Gm9694 |
predicted gene 9694 |
17304 |
0.12 |
| chr6_39380969_39382023 | 1.03 |
Rab19 |
RAB19, member RAS oncogene family |
321 |
0.84 |
| chr2_120153312_120153463 | 1.02 |
Ehd4 |
EH-domain containing 4 |
1075 |
0.49 |
| chr10_38967267_38967717 | 1.00 |
Lama4 |
laminin, alpha 4 |
1820 |
0.42 |
| chr2_77168707_77169120 | 0.98 |
Ccdc141 |
coiled-coil domain containing 141 |
1663 |
0.41 |
| chr16_43363086_43363291 | 0.98 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
140 |
0.83 |
| chr7_17060653_17061482 | 0.94 |
Hif3a |
hypoxia inducible factor 3, alpha subunit |
1317 |
0.27 |
| chr6_54330304_54330947 | 0.93 |
9130019P16Rik |
RIKEN cDNA 9130019P16 gene |
3284 |
0.22 |
| chr2_168589071_168590755 | 0.89 |
Nfatc2 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
278 |
0.93 |
| chr12_90737821_90739146 | 0.89 |
Dio2 |
deiodinase, iodothyronine, type II |
45 |
0.78 |
| chr2_118112796_118113528 | 0.88 |
Thbs1 |
thrombospondin 1 |
1286 |
0.33 |
| chr3_53657521_53658180 | 0.85 |
Frem2 |
Fras1 related extracellular matrix protein 2 |
495 |
0.76 |
| chr11_55417729_55419811 | 0.81 |
Sparc |
secreted acidic cysteine rich glycoprotein |
1128 |
0.44 |
| chr7_105738238_105738468 | 0.80 |
Rrp8 |
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
968 |
0.29 |
| chr5_116288594_116290345 | 0.80 |
Ccdc60 |
coiled-coil domain containing 60 |
484 |
0.71 |
| chr13_102903771_102904870 | 0.80 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
1466 |
0.56 |
| chr2_35463798_35464176 | 0.78 |
Ggta1 |
glycoprotein galactosyltransferase alpha 1, 3 |
756 |
0.55 |
| chr1_153279868_153280137 | 0.77 |
Gm38009 |
predicted gene, 38009 |
7188 |
0.18 |
| chr4_127311434_127313205 | 0.76 |
Gja4 |
gap junction protein, alpha 4 |
1720 |
0.26 |
| chrX_133980931_133982115 | 0.75 |
Sytl4 |
synaptotagmin-like 4 |
260 |
0.92 |
| chr5_123318053_123318557 | 0.74 |
Gm15857 |
predicted gene 15857 |
7981 |
0.1 |
| chr10_44011393_44011544 | 0.72 |
Crybg1 |
crystallin beta-gamma domain containing 1 |
7099 |
0.19 |
| chr4_14621039_14622190 | 0.72 |
Slc26a7 |
solute carrier family 26, member 7 |
55 |
0.99 |
| chr10_42581935_42584872 | 0.72 |
Nr2e1 |
nuclear receptor subfamily 2, group E, member 1 |
229 |
0.69 |
| chr9_110764325_110764587 | 0.71 |
Myl3 |
myosin, light polypeptide 3 |
810 |
0.48 |
| chr15_7272973_7273124 | 0.71 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
18280 |
0.24 |
| chr2_137107794_137108485 | 0.71 |
Jag1 |
jagged 1 |
8505 |
0.28 |
| chr5_124528520_124529412 | 0.71 |
Gm43034 |
predicted gene 43034 |
2167 |
0.16 |
| chr17_81736193_81736564 | 0.70 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
1999 |
0.44 |
| chr8_72137457_72137608 | 0.69 |
Tpm4 |
tropomyosin 4 |
1724 |
0.17 |
| chr17_48019794_48019945 | 0.69 |
Gm14871 |
predicted gene 14871 |
16297 |
0.12 |
| chr10_117235571_117236047 | 0.69 |
9530003J23Rik |
RIKEN cDNA 9530003J23 gene |
2845 |
0.16 |
| chr9_123489714_123489865 | 0.69 |
Limd1 |
LIM domains containing 1 |
9179 |
0.18 |
| chr18_42053706_42054633 | 0.68 |
Sh3rf2 |
SH3 domain containing ring finger 2 |
459 |
0.84 |
| chr4_58286744_58287250 | 0.67 |
Musk |
muscle, skeletal, receptor tyrosine kinase |
822 |
0.73 |
| chr6_88200114_88201718 | 0.66 |
Gata2 |
GATA binding protein 2 |
3 |
0.96 |
| chr2_6352474_6353269 | 0.66 |
Usp6nl |
USP6 N-terminal like |
138 |
0.96 |
| chr4_49691879_49692113 | 0.65 |
Ppp3r2 |
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
9972 |
0.18 |
| chr18_61256820_61257406 | 0.65 |
Rps2-ps10 |
ribosomal protein S2, pseudogene 10 |
2848 |
0.23 |
| chr7_128205575_128206445 | 0.62 |
Cox6a2 |
cytochrome c oxidase subunit 6A2 |
377 |
0.67 |
| chr12_118660796_118661641 | 0.61 |
Gm9267 |
predicted gene 9267 |
58541 |
0.12 |
| chr7_128003842_128005372 | 0.60 |
Trim72 |
tripartite motif-containing 72 |
229 |
0.69 |
| chr18_5027813_5027964 | 0.59 |
Svil |
supervillin |
7278 |
0.31 |
| chr7_135526475_135527168 | 0.57 |
Clrn3 |
clarin 3 |
1833 |
0.31 |
| chr7_100657972_100658903 | 0.57 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
15 |
0.96 |
| chr1_171767462_171768093 | 0.57 |
Slamf1 |
signaling lymphocytic activation molecule family member 1 |
622 |
0.59 |
| chr4_56944091_56945052 | 0.56 |
Tmem245 |
transmembrane protein 245 |
2840 |
0.22 |
| chr8_124991174_124991811 | 0.55 |
Tsnax |
translin-associated factor X |
21505 |
0.13 |
| chr1_173802084_173802692 | 0.55 |
Ifi203-ps |
interferon activated gene 203, pseudogene |
4118 |
0.16 |
| chr2_152626740_152627949 | 0.54 |
Rem1 |
rad and gem related GTP binding protein 1 |
391 |
0.68 |
| chr16_5131498_5132854 | 0.54 |
Ppl |
periplakin |
234 |
0.89 |
| chr15_80012110_80012261 | 0.54 |
Gm41361 |
predicted gene, 41361 |
146 |
0.89 |
| chr9_44726537_44726746 | 0.54 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
155 |
0.89 |
| chr7_30972587_30974196 | 0.54 |
Lsr |
lipolysis stimulated lipoprotein receptor |
24 |
0.84 |
| chr8_119437065_119437967 | 0.52 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
327 |
0.86 |
| chr5_81519288_81520108 | 0.51 |
Gm43084 |
predicted gene 43084 |
16028 |
0.29 |
| chr6_129181703_129182081 | 0.51 |
Clec2d |
C-type lectin domain family 2, member d |
1277 |
0.31 |
| chrX_135574770_135575114 | 0.51 |
Nxf7 |
nuclear RNA export factor 7 |
11191 |
0.12 |
| chr9_21797731_21798548 | 0.51 |
Kank2 |
KN motif and ankyrin repeat domains 2 |
364 |
0.77 |
| chr6_51269852_51270940 | 0.50 |
Mir148a |
microRNA 148a |
486 |
0.83 |
| chr3_107356954_107357548 | 0.50 |
Gm5279 |
predicted gene 5279 |
14296 |
0.16 |
| chr9_118975609_118975835 | 0.50 |
Ctdspl |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
45855 |
0.1 |
| chr4_57560440_57561163 | 0.50 |
Pakap |
paralemmin A kinase anchor protein |
7358 |
0.28 |
| chr2_155611238_155612364 | 0.50 |
Myh7b |
myosin, heavy chain 7B, cardiac muscle, beta |
589 |
0.53 |
| chr8_61908416_61909024 | 0.49 |
4930512H18Rik |
RIKEN cDNA 4930512H18 gene |
4340 |
0.19 |
| chr11_72409874_72410296 | 0.49 |
Smtnl2 |
smoothelin-like 2 |
1626 |
0.27 |
| chr16_16213032_16214454 | 0.49 |
Pkp2 |
plakophilin 2 |
425 |
0.85 |
| chr1_72873610_72874834 | 0.49 |
Igfbp5 |
insulin-like growth factor binding protein 5 |
53 |
0.98 |
| chr16_57551520_57552150 | 0.48 |
Filip1l |
filamin A interacting protein 1-like |
2593 |
0.35 |
| chr16_10313185_10314434 | 0.48 |
Emp2 |
epithelial membrane protein 2 |
62 |
0.97 |
| chr16_23110812_23112317 | 0.48 |
Gm24616 |
predicted gene, 24616 |
53 |
0.67 |
| chr16_92358680_92359210 | 0.48 |
Kcne1 |
potassium voltage-gated channel, Isk-related subfamily, member 1 |
71 |
0.96 |
| chr2_167542481_167542796 | 0.47 |
Snai1 |
snail family zinc finger 1 |
4443 |
0.14 |
| chr19_7557459_7558480 | 0.47 |
Plaat3 |
phospholipase A and acyltransferase 3 |
484 |
0.77 |
| chr2_59158310_59158473 | 0.46 |
Ccdc148 |
coiled-coil domain containing 148 |
2143 |
0.28 |
| chr2_4299460_4299841 | 0.46 |
Frmd4a |
FERM domain containing 4A |
868 |
0.47 |
| chr12_17324246_17325364 | 0.46 |
Atp6v1c2 |
ATPase, H+ transporting, lysosomal V1 subunit C2 |
75 |
0.96 |
| chr16_23984336_23985564 | 0.46 |
Gm37419 |
predicted gene, 37419 |
3532 |
0.2 |
| chr11_87662563_87663695 | 0.45 |
Rnf43 |
ring finger protein 43 |
42 |
0.96 |
| chr2_158145151_158146425 | 0.45 |
Tgm2 |
transglutaminase 2, C polypeptide |
578 |
0.71 |
| chr6_136660962_136662291 | 0.45 |
Plbd1 |
phospholipase B domain containing 1 |
247 |
0.91 |
| chr2_121441930_121443212 | 0.45 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
30 |
0.93 |
| chr8_106136230_106137393 | 0.45 |
Esrp2 |
epithelial splicing regulatory protein 2 |
2 |
0.77 |
| chr17_45626838_45627013 | 0.44 |
Gm25008 |
predicted gene, 25008 |
6246 |
0.1 |
| chr3_108364558_108365384 | 0.44 |
Mybphl |
myosin binding protein H-like |
60 |
0.94 |
| chr5_119681870_119683681 | 0.44 |
Tbx3 |
T-box 3 |
2526 |
0.22 |
| chr15_97049553_97050500 | 0.44 |
Slc38a4 |
solute carrier family 38, member 4 |
61 |
0.98 |
| chr9_58156326_58158409 | 0.44 |
Islr |
immunoglobulin superfamily containing leucine-rich repeat |
1178 |
0.35 |
| chr9_31130557_31131972 | 0.43 |
St14 |
suppression of tumorigenicity 14 (colon carcinoma) |
463 |
0.79 |
| chr2_174287268_174287478 | 0.43 |
Gnasas1 |
GNAS antisense RNA 1 |
1938 |
0.21 |
| chr13_113013114_113013713 | 0.43 |
Mcidas |
multiciliate differentiation and DNA synthesis associated cell cycle protein |
19568 |
0.08 |
| chr7_28370288_28370439 | 0.42 |
Plekhg2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
1145 |
0.24 |
| chr10_17797266_17797516 | 0.42 |
Txlnb |
taxilin beta |
1135 |
0.43 |
| chr4_95107063_95107214 | 0.42 |
Junos |
jun proto-oncogene, opposite strand |
8434 |
0.14 |
| chr2_140763294_140763859 | 0.42 |
Flrt3 |
fibronectin leucine rich transmembrane protein 3 |
92107 |
0.09 |
| chr16_8108271_8108713 | 0.42 |
Gm49535 |
predicted gene, 49535 |
4704 |
0.34 |
| chr11_68555954_68556965 | 0.42 |
Mfsd6l |
major facilitator superfamily domain containing 6-like |
273 |
0.89 |
| chr2_139679952_139680200 | 0.41 |
Ism1 |
isthmin 1, angiogenesis inhibitor |
1898 |
0.34 |
| chr6_33057859_33058010 | 0.41 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
2283 |
0.28 |
| chr7_89405888_89406433 | 0.41 |
Fzd4 |
frizzled class receptor 4 |
1805 |
0.24 |
| chr8_71523614_71524679 | 0.41 |
Gm7850 |
predicted gene 7850 |
572 |
0.48 |
| chr9_40877257_40878943 | 0.40 |
Bsx |
brain specific homeobox |
3973 |
0.16 |
| chr2_35132236_35133300 | 0.40 |
Cntrl |
centriolin |
510 |
0.77 |
| chr13_16018459_16019177 | 0.40 |
Inhba |
inhibin beta-A |
4343 |
0.15 |
| chr14_16572461_16573738 | 0.40 |
Rarb |
retinoic acid receptor, beta |
1946 |
0.34 |
| chr3_10366508_10367397 | 0.40 |
Chmp4c |
charged multivesicular body protein 4C |
6 |
0.96 |
| chr6_34862829_34863673 | 0.39 |
Tmem140 |
transmembrane protein 140 |
11 |
0.96 |
| chr9_61227699_61228136 | 0.39 |
Gm34105 |
predicted gene, 34105 |
21238 |
0.18 |
| chr9_21851806_21852608 | 0.39 |
Dock6 |
dedicator of cytokinesis 6 |
409 |
0.74 |
| chr16_4641802_4643186 | 0.39 |
Dnaja3 |
DnaJ heat shock protein family (Hsp40) member A3 |
2505 |
0.16 |
| chr11_120658808_120659482 | 0.38 |
Notumos |
notum palmitoleoyl-protein carboxylesterase, opposite strand |
12 |
0.9 |
| chr9_66512511_66514532 | 0.38 |
Fbxl22 |
F-box and leucine-rich repeat protein 22 |
1088 |
0.47 |
| chr10_58326224_58326780 | 0.38 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
3036 |
0.22 |
| chr8_46490914_46491682 | 0.38 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
324 |
0.87 |
| chr17_22685333_22685484 | 0.38 |
Gm9805 |
predicted gene 9805 |
2312 |
0.26 |
| chr2_119619277_119620230 | 0.37 |
Nusap1 |
nucleolar and spindle associated protein 1 |
1024 |
0.3 |
| chr18_21296515_21297047 | 0.37 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
3342 |
0.22 |
| chr15_80081033_80081744 | 0.37 |
Rpl3 |
ribosomal protein L3 |
907 |
0.27 |
| chr6_85837036_85837692 | 0.37 |
Nat8 |
N-acetyltransferase 8 (GCN5-related) |
5282 |
0.09 |
| chrX_53053916_53055220 | 0.37 |
Mir322 |
microRNA 322 |
219 |
0.66 |
| chr6_114917046_114918071 | 0.37 |
Vgll4 |
vestigial like family member 4 |
4258 |
0.26 |
| chr10_57499477_57499901 | 0.36 |
Hsf2 |
heat shock factor 2 |
685 |
0.68 |
| chr1_72960515_72960666 | 0.36 |
1700027A15Rik |
RIKEN cDNA 1700027A15 gene |
24254 |
0.2 |
| chr1_173941834_173942320 | 0.36 |
Ifi203 |
interferon activated gene 203 |
377 |
0.8 |
| chr10_56377345_56379375 | 0.36 |
Gja1 |
gap junction protein, alpha 1 |
82 |
0.97 |
| chrX_102187053_102187918 | 0.36 |
Rps4x |
ribosomal protein S4, X-linked |
748 |
0.53 |
| chr11_101891234_101892171 | 0.36 |
Meox1 |
mesenchyme homeobox 1 |
2672 |
0.18 |
| chr12_69447750_69448552 | 0.36 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
19241 |
0.14 |
| chr13_109229388_109230388 | 0.36 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
30766 |
0.25 |
| chr10_111957572_111957832 | 0.36 |
Krr1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
14962 |
0.16 |
| chr14_34375555_34377243 | 0.35 |
Mmrn2 |
multimerin 2 |
678 |
0.52 |
| chr8_70765697_70766944 | 0.35 |
6330537M06Rik |
RIKEN cDNA 6330537M06 gene |
259 |
0.52 |
| chr17_70997235_70997648 | 0.35 |
Myl12a |
myosin, light chain 12A, regulatory, non-sarcomeric |
134 |
0.92 |
| chr5_35391428_35392811 | 0.35 |
4930442P19Rik |
RIKEN cDNA 4930442P19 gene |
207 |
0.9 |
| chr8_69992523_69992674 | 0.35 |
Gatad2a |
GATA zinc finger domain containing 2A |
3635 |
0.15 |
| chr1_86580854_86581005 | 0.35 |
Gm37152 |
predicted gene, 37152 |
871 |
0.4 |
| chr9_102643756_102644501 | 0.35 |
Anapc13 |
anaphase promoting complex subunit 13 |
15616 |
0.12 |
| chr19_32956612_32957351 | 0.34 |
Gm36860 |
predicted gene, 36860 |
23082 |
0.24 |
| chr3_8594088_8594846 | 0.34 |
Gm18822 |
predicted gene, 18822 |
22037 |
0.15 |
| chr1_157413776_157413976 | 0.34 |
2810025M15Rik |
RIKEN cDNA 2810025M15 gene |
898 |
0.4 |
| chr8_61273636_61273787 | 0.34 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
15161 |
0.2 |
| chr10_69617478_69618016 | 0.34 |
Gm46231 |
predicted gene, 46231 |
51386 |
0.16 |
| chr2_115512707_115513160 | 0.34 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
726 |
0.75 |
| chr9_63749888_63750053 | 0.34 |
Smad3 |
SMAD family member 3 |
1107 |
0.57 |
| chr18_37821959_37823152 | 0.34 |
Gm38182 |
predicted gene, 38182 |
1324 |
0.18 |
| chr11_77798312_77798528 | 0.33 |
Myo18a |
myosin XVIIIA |
2878 |
0.21 |
| chr18_82821622_82822615 | 0.33 |
4930445N18Rik |
RIKEN cDNA 4930445N18 gene |
33776 |
0.11 |
| chr5_96983338_96983489 | 0.33 |
Gm9484 |
predicted gene 9484 |
13951 |
0.12 |
| chr15_85734856_85735435 | 0.33 |
Ppara |
peroxisome proliferator activated receptor alpha |
162 |
0.94 |
| chr2_33218132_33218667 | 0.33 |
Angptl2 |
angiopoietin-like 2 |
2282 |
0.25 |
| chr17_85607592_85608395 | 0.33 |
Six3 |
sine oculis-related homeobox 3 |
5615 |
0.15 |
| chr4_41706867_41707261 | 0.33 |
Rpp25l |
ribonuclease P/MRP 25 subunit-like |
6470 |
0.09 |
| chr9_40324933_40325121 | 0.33 |
1700110K17Rik |
RIKEN cDNA 1700110K17 gene |
1601 |
0.25 |
| chr12_25092495_25093804 | 0.33 |
Id2 |
inhibitor of DNA binding 2 |
2938 |
0.22 |
| chr7_15963733_15964484 | 0.33 |
Ehd2 |
EH-domain containing 2 |
3367 |
0.13 |
| chr4_45965010_45965225 | 0.33 |
Tdrd7 |
tudor domain containing 7 |
217 |
0.94 |
| chr1_136007228_136008019 | 0.33 |
Tmem9 |
transmembrane protein 9 |
527 |
0.63 |
| chr1_118481967_118482873 | 0.32 |
Clasp1 |
CLIP associating protein 1 |
381 |
0.8 |
| chr2_35460032_35460183 | 0.32 |
Ggta1 |
glycoprotein galactosyltransferase alpha 1, 3 |
1142 |
0.39 |
| chr7_100926314_100930096 | 0.32 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
3902 |
0.17 |
| chr2_104848924_104850114 | 0.32 |
Prrg4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
26 |
0.97 |
| chr9_50752994_50754649 | 0.32 |
Cryab |
crystallin, alpha B |
574 |
0.54 |
| chr11_88516893_88517089 | 0.31 |
Msi2 |
musashi RNA-binding protein 2 |
73156 |
0.09 |
| chr11_72301272_72302549 | 0.31 |
Xaf1 |
XIAP associated factor 1 |
245 |
0.87 |
| chr1_161713119_161713350 | 0.31 |
Gm37638 |
predicted gene, 37638 |
471 |
0.79 |
| chr10_78399155_78399306 | 0.31 |
Rrp1 |
ribosomal RNA processing 1 homolog (S. cerevisiae) |
4386 |
0.09 |
| chr9_42275689_42275840 | 0.31 |
Gm36435 |
predicted gene, 36435 |
4114 |
0.2 |
| chr12_3806578_3808221 | 0.31 |
Dnmt3a |
DNA methyltransferase 3A |
239 |
0.92 |
| chr5_122102345_122102815 | 0.30 |
Myl2 |
myosin, light polypeptide 2, regulatory, cardiac, slow |
599 |
0.66 |
| chr13_112425188_112425430 | 0.30 |
Gm48876 |
predicted gene, 48876 |
1031 |
0.45 |
| chr5_114512883_114513065 | 0.30 |
Gm13790 |
predicted gene 13790 |
38315 |
0.11 |
| chr4_48663556_48664483 | 0.30 |
Cavin4 |
caveolae associated 4 |
505 |
0.78 |
| chr1_78009156_78009351 | 0.30 |
Gm28387 |
predicted gene 28387 |
36877 |
0.21 |
| chr9_7983652_7983803 | 0.30 |
Gm20416 |
predicted gene 20416 |
17889 |
0.14 |
| chr17_44188075_44189373 | 0.30 |
Clic5 |
chloride intracellular channel 5 |
140 |
0.98 |
| chr3_128239796_128239947 | 0.30 |
D030025E07Rik |
RIKEN cDNA D030025E07 gene |
8265 |
0.25 |
| chr2_105129293_105129444 | 0.30 |
Wt1 |
Wilms tumor 1 homolog |
1432 |
0.37 |
| chr5_149312765_149313642 | 0.30 |
Gm19719 |
predicted gene, 19719 |
25335 |
0.09 |
| chr11_76622309_76623770 | 0.30 |
Abr |
active BCR-related gene |
519 |
0.78 |
| chr13_34650593_34650744 | 0.30 |
Pxdc1 |
PX domain containing 1 |
2013 |
0.21 |
| chr3_133235248_133235399 | 0.30 |
Arhgef38 |
Rho guanine nucleotide exchange factor (GEF) 38 |
374 |
0.89 |
| chr18_44383580_44383731 | 0.29 |
Dcp2 |
decapping mRNA 2 |
3128 |
0.28 |
| chr8_118162236_118162415 | 0.29 |
Gm25200 |
predicted gene, 25200 |
41371 |
0.2 |
| chr2_46442111_46442262 | 0.29 |
Gm13470 |
predicted gene 13470 |
524 |
0.85 |
| chr3_123235460_123236700 | 0.29 |
Synpo2 |
synaptopodin 2 |
16 |
0.97 |
| chr10_120169087_120169245 | 0.29 |
Gm47009 |
predicted gene, 47009 |
517 |
0.66 |
| chr11_83855419_83855971 | 0.29 |
Hnf1b |
HNF1 homeobox B |
2735 |
0.21 |
| chr2_33753588_33754077 | 0.29 |
Mvb12b |
multivesicular body subunit 12B |
34927 |
0.14 |
| chr1_173879500_173880236 | 0.29 |
Mndal |
myeloid nuclear differentiation antigen like |
243 |
0.89 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.7 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.3 | 0.9 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 0.9 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.7 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.0 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.2 | 0.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.7 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.1 | 0.5 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.3 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 0.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.5 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.2 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 0.2 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.2 | GO:0003164 | His-Purkinje system development(GO:0003164) bundle of His development(GO:0003166) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.5 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.9 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.0 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.6 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0061550 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cranial ganglion development(GO:0061550) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.6 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0055119 | relaxation of cardiac muscle(GO:0055119) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:1902218 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.0 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 3.2 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 0.5 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.6 | GO:0043905 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0034819 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |