Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
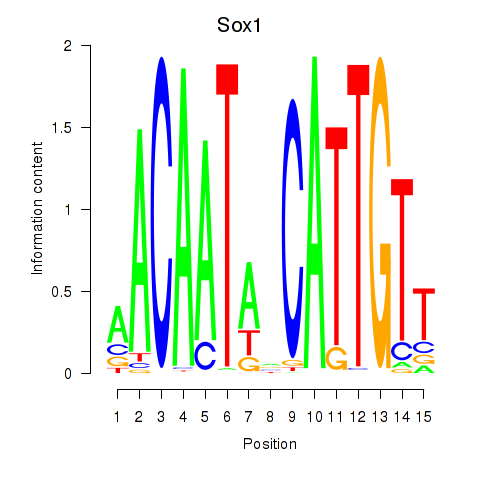
Results for Sox1
Z-value: 0.98
Transcription factors associated with Sox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox1
|
ENSMUSG00000096014.1 | SRY (sex determining region Y)-box 1 |
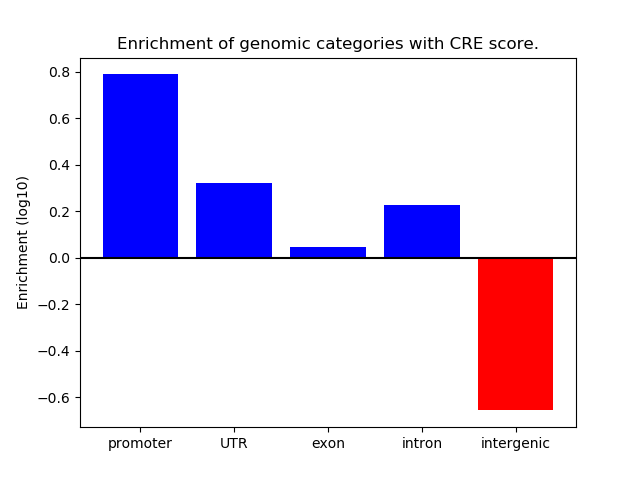
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_12393247_12393480 | Sox1 | 1932 | 0.221782 | 0.31 | 1.8e-02 | Click! |
| chr8_12394789_12395390 | Sox1 | 206 | 0.880732 | -0.27 | 3.4e-02 | Click! |
| chr8_12395440_12396127 | Sox1 | 488 | 0.504407 | 0.19 | 1.5e-01 | Click! |
| chr8_12394043_12394245 | Sox1 | 1151 | 0.353540 | 0.15 | 2.5e-01 | Click! |
| chr8_12394304_12394455 | Sox1 | 916 | 0.433980 | 0.11 | 3.9e-01 | Click! |
Activity of the Sox1 motif across conditions
Conditions sorted by the z-value of the Sox1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
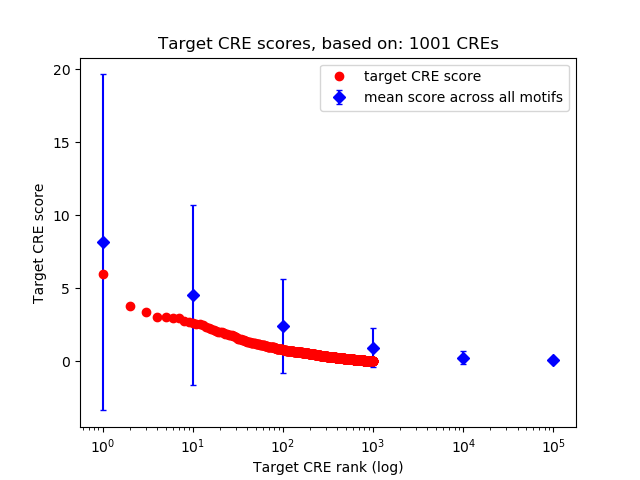
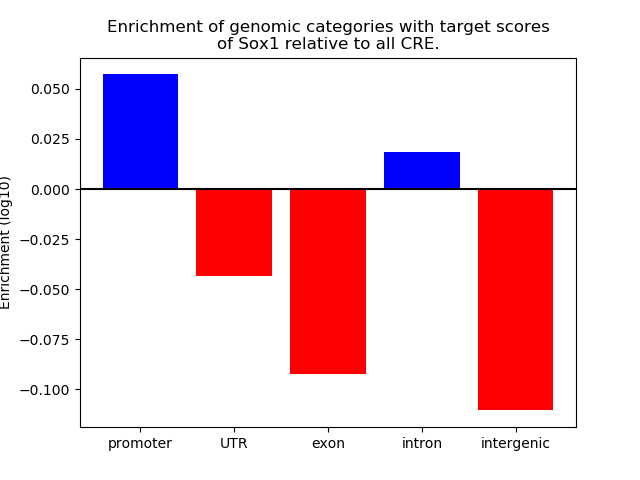
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_17750183_17750783 | 6.01 |
St7 |
suppression of tumorigenicity 7 |
1091 |
0.38 |
| chr2_181313043_181314281 | 3.79 |
Stmn3 |
stathmin-like 3 |
838 |
0.42 |
| chr13_99513069_99513554 | 3.37 |
Map1b |
microtubule-associated protein 1B |
3207 |
0.2 |
| chr6_55680133_55680881 | 3.04 |
Neurod6 |
neurogenic differentiation 6 |
756 |
0.69 |
| chrX_23284413_23285126 | 3.01 |
Klhl13 |
kelch-like 13 |
60 |
0.99 |
| chr8_14382368_14383445 | 2.98 |
Dlgap2 |
DLG associated protein 2 |
910 |
0.66 |
| chr4_110282527_110283235 | 2.95 |
Elavl4 |
ELAV like RNA binding protein 4 |
3735 |
0.36 |
| chr7_54866491_54866642 | 2.76 |
Gm44895 |
predicted gene 44895 |
9548 |
0.19 |
| chr7_51620788_51621096 | 2.70 |
Slc17a6 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
1064 |
0.54 |
| chr6_8948312_8949000 | 2.61 |
Nxph1 |
neurexophilin 1 |
42 |
0.99 |
| chr4_24429901_24430719 | 2.54 |
Gm27243 |
predicted gene 27243 |
580 |
0.79 |
| chr13_62967411_62968273 | 2.53 |
Aopep |
aminopeptidase O |
2949 |
0.22 |
| chr3_68573207_68574269 | 2.45 |
Schip1 |
schwannomin interacting protein 1 |
1493 |
0.45 |
| chrX_84076569_84077653 | 2.36 |
Dmd |
dystrophin, muscular dystrophy |
462 |
0.87 |
| chr14_65423052_65425451 | 2.26 |
Pnoc |
prepronociceptin |
909 |
0.6 |
| chr1_120118110_120118594 | 2.23 |
Dbi |
diazepam binding inhibitor |
1786 |
0.31 |
| chr11_41997931_41998166 | 2.17 |
Gabrg2 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
2308 |
0.39 |
| chr9_41377643_41378358 | 2.10 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
1439 |
0.4 |
| chr5_88587035_88587274 | 2.01 |
Rufy3 |
RUN and FYVE domain containing 3 |
3360 |
0.21 |
| chr12_52702153_52702917 | 2.00 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
3152 |
0.28 |
| chr7_62366195_62366527 | 1.99 |
Magel2 |
melanoma antigen, family L, 2 |
10649 |
0.18 |
| chr2_52557337_52558561 | 1.96 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
611 |
0.74 |
| chr19_6499251_6500132 | 1.87 |
Nrxn2 |
neurexin II |
1856 |
0.23 |
| chr12_24976343_24976494 | 1.84 |
Gm36236 |
predicted gene, 36236 |
864 |
0.51 |
| chr11_34102897_34103093 | 1.79 |
4930469K13Rik |
RIKEN cDNA 4930469K13 gene |
4441 |
0.22 |
| chr5_111425949_111427685 | 1.79 |
Gm43119 |
predicted gene 43119 |
3228 |
0.22 |
| chr8_54956010_54956394 | 1.78 |
Gpm6a |
glycoprotein m6a |
1359 |
0.38 |
| chr10_110453550_110454045 | 1.74 |
Nav3 |
neuron navigator 3 |
2407 |
0.34 |
| chr4_82507895_82509049 | 1.71 |
Gm11266 |
predicted gene 11266 |
456 |
0.82 |
| chr1_20892892_20893201 | 1.64 |
Paqr8 |
progestin and adipoQ receptor family member VIII |
2440 |
0.18 |
| chr12_74285521_74285936 | 1.58 |
1700086L19Rik |
RIKEN cDNA 1700086L19 gene |
1439 |
0.35 |
| chr6_123296087_123296238 | 1.52 |
Clec4e |
C-type lectin domain family 4, member e |
6292 |
0.15 |
| chr16_42336507_42337221 | 1.50 |
Gap43 |
growth associated protein 43 |
3787 |
0.3 |
| chr3_119064938_119065089 | 1.50 |
Gm43410 |
predicted gene 43410 |
196769 |
0.03 |
| chr17_78508180_78509392 | 1.48 |
Vit |
vitrin |
614 |
0.7 |
| chr7_70372558_70372768 | 1.45 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
5928 |
0.13 |
| chrX_158924759_158924985 | 1.43 |
Gm5764 |
predicted gene 5764 |
91437 |
0.09 |
| chr5_51495096_51495739 | 1.41 |
Gm42615 |
predicted gene 42615 |
2501 |
0.23 |
| chr5_58747985_58748136 | 1.36 |
Gm43394 |
predicted gene 43394 |
252933 |
0.02 |
| chr4_24429141_24429555 | 1.34 |
Gm27243 |
predicted gene 27243 |
1542 |
0.44 |
| chr1_81077232_81078427 | 1.34 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
246 |
0.96 |
| chr11_87985633_87986121 | 1.34 |
Dynll2 |
dynein light chain LC8-type 2 |
708 |
0.59 |
| chr16_33686218_33687745 | 1.29 |
Heg1 |
heart development protein with EGF-like domains 1 |
2436 |
0.35 |
| chr10_115819869_115820089 | 1.27 |
Tspan8 |
tetraspanin 8 |
2695 |
0.36 |
| chr14_90028075_90028226 | 1.27 |
Gm48941 |
predicted gene, 48941 |
57742 |
0.15 |
| chr5_104608353_104608504 | 1.27 |
Thoc2l |
THO complex subunit 2-like |
67245 |
0.1 |
| chr7_76889140_76890193 | 1.24 |
Gm45210 |
predicted gene 45210 |
189949 |
0.03 |
| chr1_168676421_168677250 | 1.23 |
1700063I16Rik |
RIKEN cDNA 1700063I16 gene |
953 |
0.73 |
| chr4_27987811_27988244 | 1.22 |
Tpm3-rs2 |
tropomyosin 3, related sequence 2 |
8878 |
0.29 |
| chr18_60925301_60926809 | 1.22 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
330 |
0.84 |
| chr16_49572852_49573281 | 1.20 |
Ift57 |
intraflagellar transport 57 |
126167 |
0.05 |
| chr6_81660825_81661326 | 1.20 |
Gm26264 |
predicted gene, 26264 |
22764 |
0.21 |
| chr14_65260243_65260419 | 1.18 |
Fzd3 |
frizzled class receptor 3 |
2132 |
0.26 |
| chrX_106186521_106186932 | 1.17 |
Pgk1 |
phosphoglycerate kinase 1 |
374 |
0.82 |
| chr16_72030928_72031079 | 1.16 |
Gm49667 |
predicted gene, 49667 |
151959 |
0.04 |
| chr7_36480723_36480874 | 1.15 |
Gm25247 |
predicted gene, 25247 |
50187 |
0.15 |
| chr5_28206868_28207439 | 1.14 |
Cnpy1 |
canopy FGF signaling regulator 1 |
2869 |
0.27 |
| chr8_9767652_9767828 | 1.14 |
Fam155a |
family with sequence similarity 155, member A |
2364 |
0.22 |
| chr1_63275593_63275905 | 1.13 |
Gm24513 |
predicted gene, 24513 |
2386 |
0.15 |
| chr15_85578626_85579014 | 1.13 |
Wnt7b |
wingless-type MMTV integration site family, member 7B |
750 |
0.54 |
| chrX_119427268_119427419 | 1.13 |
Gm14926 |
predicted gene 14926 |
14091 |
0.29 |
| chr10_40885530_40885990 | 1.13 |
Wasf1 |
WAS protein family, member 1 |
1933 |
0.32 |
| chr6_65676533_65676711 | 1.04 |
Ndnf |
neuron-derived neurotrophic factor |
5032 |
0.25 |
| chr9_85525791_85525942 | 1.03 |
Gm48834 |
predicted gene, 48834 |
27899 |
0.18 |
| chr4_57637702_57639097 | 1.03 |
Pakap |
paralemmin A kinase anchor protein |
424 |
0.89 |
| chr5_67746354_67746505 | 1.03 |
Gm42737 |
predicted gene 42737 |
12901 |
0.16 |
| chr3_87566316_87566693 | 1.01 |
ETV3L |
ets variant 3-like |
15255 |
0.14 |
| chr9_101862248_101863069 | 1.00 |
Gm29521 |
predicted gene 29521 |
169 |
0.89 |
| chr5_125267591_125268229 | 1.00 |
Gm32585 |
predicted gene, 32585 |
4905 |
0.19 |
| chr1_152764641_152764934 | 0.99 |
Rgl1 |
ral guanine nucleotide dissociation stimulator,-like 1 |
1518 |
0.25 |
| chr15_44706791_44706942 | 0.99 |
Sybu |
syntabulin (syntaxin-interacting) |
40922 |
0.15 |
| chr17_80847108_80847654 | 0.98 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
49412 |
0.14 |
| chr14_113319455_113319606 | 0.97 |
Tpm3-rs7 |
tropomyosin 3, related sequence 7 |
4922 |
0.35 |
| chr13_83984481_83985348 | 0.97 |
Gm4241 |
predicted gene 4241 |
3077 |
0.26 |
| chr1_106176006_106176160 | 0.97 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
4331 |
0.2 |
| chr2_136058799_136059736 | 0.96 |
Lamp5 |
lysosomal-associated membrane protein family, member 5 |
164 |
0.96 |
| chr5_116586161_116587058 | 0.96 |
Srrm4 |
serine/arginine repetitive matrix 4 |
5208 |
0.2 |
| chr19_37176789_37177437 | 0.96 |
Cpeb3 |
cytoplasmic polyadenylation element binding protein 3 |
904 |
0.43 |
| chr14_55055914_55056065 | 0.95 |
Gm20687 |
predicted gene 20687 |
496 |
0.57 |
| chr10_106469534_106470969 | 0.92 |
Ppfia2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
88 |
0.97 |
| chr13_104393590_104393741 | 0.92 |
Adamts6 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
86318 |
0.09 |
| chr14_79533605_79533836 | 0.90 |
Elf1 |
E74-like factor 1 |
18022 |
0.15 |
| chrX_56736306_56736457 | 0.89 |
Fhl1 |
four and a half LIM domains 1 |
4252 |
0.19 |
| chrX_105390628_105392456 | 0.88 |
5330434G04Rik |
RIKEN cDNA 5330434G04 gene |
212 |
0.93 |
| chr14_61174181_61174611 | 0.88 |
Sacs |
sacsin |
1406 |
0.46 |
| chr17_72601322_72601675 | 0.87 |
Alk |
anaplastic lymphoma kinase |
2329 |
0.39 |
| chr1_186278091_186278855 | 0.87 |
Gm37491 |
predicted gene, 37491 |
68842 |
0.11 |
| chr1_88698575_88698781 | 0.86 |
Arl4c |
ADP-ribosylation factor-like 4C |
3198 |
0.22 |
| chr19_40510615_40511718 | 0.86 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
2507 |
0.27 |
| chr16_6841576_6842071 | 0.85 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
32601 |
0.26 |
| chr7_97842700_97844158 | 0.84 |
Pak1 |
p21 (RAC1) activated kinase 1 |
494 |
0.83 |
| chr4_57637428_57637579 | 0.84 |
Pakap |
paralemmin A kinase anchor protein |
320 |
0.93 |
| chr7_126700976_126702469 | 0.83 |
Coro1a |
coronin, actin binding protein 1A |
373 |
0.64 |
| chr12_108118899_108119566 | 0.83 |
Setd3 |
SET domain containing 3 |
4149 |
0.29 |
| chr2_22775769_22775920 | 0.82 |
Apbb1ip |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
1399 |
0.36 |
| chr3_34958948_34959456 | 0.82 |
Mir6378 |
microRNA 6378 |
36551 |
0.2 |
| chr16_45407023_45407415 | 0.81 |
Cd200 |
CD200 antigen |
1645 |
0.32 |
| chr3_114904046_114905354 | 0.81 |
Olfm3 |
olfactomedin 3 |
65 |
0.98 |
| chr6_23877057_23877231 | 0.80 |
Cadps2 |
Ca2+-dependent activator protein for secretion 2 |
37723 |
0.2 |
| chrX_101642899_101643454 | 0.80 |
Ogt |
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) |
3036 |
0.21 |
| chr13_54748886_54749162 | 0.79 |
Gprin1 |
G protein-regulated inducer of neurite outgrowth 1 |
645 |
0.59 |
| chr8_83167299_83167462 | 0.78 |
Gm10645 |
predicted gene 10645 |
1210 |
0.39 |
| chr5_118618819_118619232 | 0.78 |
Gm43275 |
predicted gene 43275 |
731 |
0.61 |
| chr10_84754760_84756248 | 0.76 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
558 |
0.8 |
| chr4_35227017_35227268 | 0.76 |
C9orf72 |
C9orf72, member of C9orf72-SMCR8 complex |
967 |
0.42 |
| chr7_30292635_30292786 | 0.75 |
Clip3 |
CAP-GLY domain containing linker protein 3 |
752 |
0.37 |
| chr11_80479429_80480178 | 0.74 |
Cdk5r1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
2747 |
0.26 |
| chr6_42265062_42266248 | 0.74 |
Casp2 |
caspase 2 |
535 |
0.62 |
| chr9_124276429_124276861 | 0.74 |
Gm39469 |
predicted gene, 39469 |
5494 |
0.18 |
| chr12_103046852_103047197 | 0.73 |
Unc79 |
unc-79 homolog |
12055 |
0.18 |
| chr16_85169623_85170249 | 0.73 |
Gm27295 |
predicted gene, 27295 |
1087 |
0.49 |
| chr1_45924247_45924747 | 0.73 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
765 |
0.56 |
| chr3_149247042_149247837 | 0.73 |
Gm10287 |
predicted gene 10287 |
21694 |
0.18 |
| chr8_110357180_110357331 | 0.72 |
Gm23627 |
predicted gene, 23627 |
64212 |
0.1 |
| chr13_69730170_69730425 | 0.72 |
Ube2ql1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
9592 |
0.12 |
| chr9_90690248_90690460 | 0.71 |
Gm2497 |
predicted gene 2497 |
43170 |
0.14 |
| chr11_120647315_120648501 | 0.71 |
Myadml2 |
myeloid-associated differentiation marker-like 2 |
202 |
0.49 |
| chr4_109342938_109343450 | 0.71 |
Eps15 |
epidermal growth factor receptor pathway substrate 15 |
59 |
0.97 |
| chrX_51219469_51219896 | 0.71 |
Gm5388 |
predicted gene 5388 |
311 |
0.62 |
| chr8_70486586_70487634 | 0.70 |
Tmem59l |
transmembrane protein 59-like |
214 |
0.81 |
| chr7_128331239_128331398 | 0.70 |
Gm44650 |
predicted gene 44650 |
1510 |
0.25 |
| chr10_102355438_102355589 | 0.70 |
Mgat4c |
MGAT4 family, member C |
16493 |
0.18 |
| chr4_33929535_33929891 | 0.70 |
Cnr1 |
cannabinoid receptor 1 (brain) |
3511 |
0.32 |
| chr18_86601591_86601919 | 0.69 |
Gm50386 |
predicted gene, 50386 |
216 |
0.96 |
| chr1_63275050_63275219 | 0.69 |
Gm24513 |
predicted gene, 24513 |
1771 |
0.18 |
| chr4_43575507_43576311 | 0.69 |
Gba2 |
glucosidase beta 2 |
1855 |
0.14 |
| chr6_87528586_87528758 | 0.69 |
Arhgap25 |
Rho GTPase activating protein 25 |
4563 |
0.17 |
| chr11_22975402_22975952 | 0.69 |
Zrsr1 |
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
2449 |
0.18 |
| chr5_114569026_114570120 | 0.68 |
Fam222a |
family with sequence similarity 222, member A |
1556 |
0.35 |
| chr4_92562847_92562998 | 0.67 |
Gm12637 |
predicted gene 12637 |
34371 |
0.24 |
| chr2_165878704_165881030 | 0.67 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
35 |
0.65 |
| chr12_84175369_84176722 | 0.67 |
Gm19327 |
predicted gene, 19327 |
11761 |
0.1 |
| chr3_8662827_8663019 | 0.67 |
Hey1 |
hairy/enhancer-of-split related with YRPW motif 1 |
3702 |
0.17 |
| chr1_111746995_111747146 | 0.66 |
Gm8173 |
predicted gene 8173 |
60705 |
0.13 |
| chr14_104641438_104641957 | 0.66 |
D130009I18Rik |
RIKEN cDNA D130009I18 gene |
2553 |
0.3 |
| chr11_30022183_30022992 | 0.66 |
Eml6 |
echinoderm microtubule associated protein like 6 |
3446 |
0.27 |
| chr6_142961825_142962306 | 0.65 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
2076 |
0.25 |
| chr9_47534660_47535132 | 0.65 |
Cadm1 |
cell adhesion molecule 1 |
4523 |
0.22 |
| chr16_3267024_3267175 | 0.65 |
Gm23215 |
predicted gene, 23215 |
17515 |
0.15 |
| chr14_105499294_105499968 | 0.65 |
4930449E01Rik |
RIKEN cDNA 4930449E01 gene |
843 |
0.63 |
| chr5_66627738_66628132 | 0.65 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
9151 |
0.15 |
| chr15_25757864_25758641 | 0.65 |
Myo10 |
myosin X |
513 |
0.83 |
| chr10_19459107_19459447 | 0.64 |
Gm33104 |
predicted gene, 33104 |
27438 |
0.19 |
| chr2_83814030_83814462 | 0.64 |
Fam171b |
family with sequence similarity 171, member B |
1610 |
0.34 |
| chr18_68972113_68972264 | 0.64 |
4930546C10Rik |
RIKEN cDNA 4930546C10 gene |
20660 |
0.22 |
| chr2_51144318_51145477 | 0.64 |
Rnd3 |
Rho family GTPase 3 |
4197 |
0.29 |
| chr15_99704098_99705242 | 0.63 |
Gm34939 |
predicted gene, 34939 |
694 |
0.3 |
| chr10_36509078_36510052 | 0.63 |
Hs3st5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
2158 |
0.44 |
| chr5_118726229_118726380 | 0.63 |
Gm43051 |
predicted gene 43051 |
18902 |
0.16 |
| chrX_114473948_114474959 | 0.63 |
Klhl4 |
kelch-like 4 |
120 |
0.97 |
| chr7_27861771_27862095 | 0.63 |
Zfp607a |
zinc finger protein 607A |
624 |
0.6 |
| chr17_21257879_21258171 | 0.63 |
Vmn1r235 |
vomeronasal 1 receptor 235 |
1614 |
0.2 |
| chr4_53632665_53633308 | 0.62 |
Fsd1l |
fibronectin type III and SPRY domain containing 1-like |
1270 |
0.45 |
| chr3_90258630_90258781 | 0.62 |
Crtc2 |
CREB regulated transcription coactivator 2 |
260 |
0.8 |
| chr1_153652756_153654124 | 0.62 |
Rgs8 |
regulator of G-protein signaling 8 |
415 |
0.8 |
| chr15_61102623_61102978 | 0.62 |
Gm38563 |
predicted gene, 38563 |
55069 |
0.14 |
| chr18_16753999_16754150 | 0.61 |
Gm15485 |
predicted gene 15485 |
25341 |
0.2 |
| chr8_45629766_45630749 | 0.61 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
2056 |
0.32 |
| chr3_133309865_133310159 | 0.61 |
Gm43254 |
predicted gene 43254 |
4 |
0.58 |
| chr12_49390931_49392462 | 0.61 |
3110039M20Rik |
RIKEN cDNA 3110039M20 gene |
1037 |
0.42 |
| chr5_4100830_4101020 | 0.61 |
Cyp51 |
cytochrome P450, family 51 |
2456 |
0.29 |
| chr1_139079212_139079368 | 0.61 |
Dennd1b |
DENN/MADD domain containing 1B |
6634 |
0.13 |
| chr12_101913750_101913901 | 0.60 |
Trip11 |
thyroid hormone receptor interactor 11 |
558 |
0.43 |
| chr8_84769068_84769679 | 0.60 |
Nfix |
nuclear factor I/X |
4023 |
0.13 |
| chr11_31684159_31684376 | 0.59 |
Bod1 |
biorientation of chromosomes in cell division 1 |
12382 |
0.22 |
| chr13_44839841_44841006 | 0.59 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
268 |
0.91 |
| chrX_160991882_160992085 | 0.59 |
Cdkl5 |
cyclin-dependent kinase-like 5 |
2721 |
0.36 |
| chrX_95195777_95197048 | 0.58 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
40 |
0.98 |
| chr18_69579071_69579222 | 0.58 |
Tcf4 |
transcription factor 4 |
13842 |
0.24 |
| chr8_16114826_16115241 | 0.58 |
Mir3106 |
microRNA 3106 |
53805 |
0.16 |
| chr13_77128205_77128356 | 0.57 |
Slf1 |
SMC5-SMC6 complex localization factor 1 |
2996 |
0.28 |
| chr19_53359518_53360664 | 0.57 |
Mxi1 |
MAX interactor 1, dimerization protein |
9932 |
0.13 |
| chr4_98094219_98094802 | 0.57 |
Gm12691 |
predicted gene 12691 |
52089 |
0.16 |
| chr2_64096278_64096678 | 0.56 |
Fign |
fidgetin |
1510 |
0.59 |
| chr17_23075779_23075930 | 0.56 |
Gm18397 |
predicted gene, 18397 |
10804 |
0.13 |
| chr10_110455062_110456014 | 0.56 |
Nav3 |
neuron navigator 3 |
666 |
0.75 |
| chrX_94507695_94508196 | 0.56 |
Maged1 |
melanoma antigen, family D, 1 |
29619 |
0.1 |
| chr12_71533269_71533420 | 0.56 |
4930404H11Rik |
RIKEN cDNA 4930404H11 gene |
7263 |
0.24 |
| chr5_21737191_21737965 | 0.56 |
Pmpcb |
peptidase (mitochondrial processing) beta |
437 |
0.76 |
| chr9_39074231_39074913 | 0.56 |
Gm8543 |
predicted gene 8543 |
470 |
0.49 |
| chr18_34505600_34505795 | 0.55 |
Fam13b |
family with sequence similarity 13, member B |
1107 |
0.43 |
| chr1_136342616_136342801 | 0.55 |
Camsap2 |
calmodulin regulated spectrin-associated protein family, member 2 |
2990 |
0.22 |
| chr14_122484799_122486138 | 0.55 |
Gm10837 |
predicted gene 10837 |
5118 |
0.12 |
| chr3_127159846_127160676 | 0.55 |
Gm16958 |
predicted gene, 16958 |
1043 |
0.4 |
| chr10_81656963_81657268 | 0.55 |
Gm10778 |
predicted gene 10778 |
4883 |
0.09 |
| chr2_38621764_38621915 | 0.54 |
Gm13586 |
predicted gene 13586 |
962 |
0.44 |
| chr10_109830543_109831446 | 0.54 |
Nav3 |
neuron navigator 3 |
2227 |
0.45 |
| chr17_34602634_34603562 | 0.54 |
Rnf5 |
ring finger protein 5 |
278 |
0.66 |
| chr18_37005167_37006150 | 0.54 |
Pcdha11 |
protocadherin alpha 11 |
455 |
0.55 |
| chr5_118666380_118667254 | 0.53 |
Gm43052 |
predicted gene 43052 |
20801 |
0.14 |
| chr4_152263958_152264463 | 0.53 |
Gpr153 |
G protein-coupled receptor 153 |
10022 |
0.12 |
| chr2_105675959_105678109 | 0.53 |
Pax6 |
paired box 6 |
905 |
0.54 |
| chr7_19932934_19933834 | 0.53 |
Igsf23 |
immunoglobulin superfamily, member 23 |
11514 |
0.07 |
| chr3_134242487_134243044 | 0.52 |
Gm26691 |
predicted gene, 26691 |
2124 |
0.19 |
| chr8_54964691_54964995 | 0.52 |
Gm45263 |
predicted gene 45263 |
5024 |
0.18 |
| chr16_62480350_62480501 | 0.52 |
CT010569.1 |
NADH dehydrogenase 5, mitochondrial (mt-Nd5) pseudogene |
3524 |
0.34 |
| chr14_75963198_75963625 | 0.52 |
Kctd4 |
potassium channel tetramerisation domain containing 4 |
8402 |
0.18 |
| chr5_116209233_116209384 | 0.52 |
Gm14507 |
predicted gene 14507 |
34398 |
0.13 |
| chr18_12938034_12938774 | 0.52 |
Osbpl1a |
oxysterol binding protein-like 1A |
3373 |
0.23 |
| chr1_52565908_52566087 | 0.51 |
Gm28178 |
predicted gene 28178 |
14294 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 0.8 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.2 | 1.0 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.2 | 0.7 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 0.7 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 0.6 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.2 | 1.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.5 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 0.5 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.4 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 2.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.4 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.4 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.3 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 1.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 1.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.7 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.3 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.2 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.1 | 0.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.3 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.1 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.2 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.6 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.3 | GO:0098597 | observational learning(GO:0098597) |
| 0.0 | 0.5 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.2 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.3 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 2.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.3 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.0 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.0 | GO:0090494 | catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0007296 | vitellogenesis(GO:0007296) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 3.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 2.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 1.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0034549 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 4.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.0 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.3 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |