Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
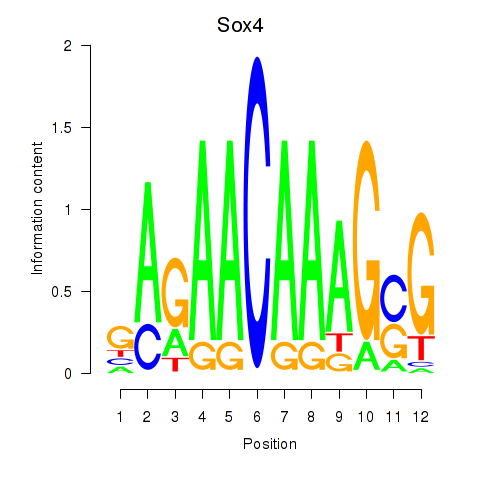
Results for Sox4
Z-value: 0.95
Transcription factors associated with Sox4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox4
|
ENSMUSG00000076431.4 | SRY (sex determining region Y)-box 4 |
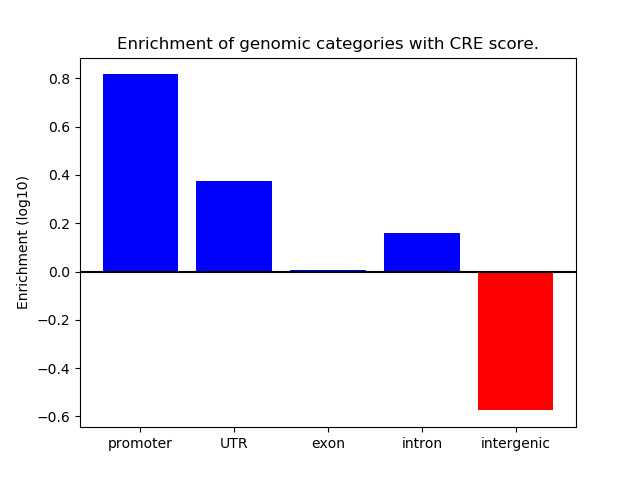
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28948063_28948214 | Sox4 | 5575 | 0.230495 | 0.62 | 1.5e-07 | Click! |
| chr13_28947811_28947962 | Sox4 | 5827 | 0.228229 | 0.53 | 1.6e-05 | Click! |
| chr13_28948354_28948958 | Sox4 | 5057 | 0.236018 | 0.52 | 1.9e-05 | Click! |
| chr13_28949179_28949330 | Sox4 | 4459 | 0.244444 | 0.46 | 2.4e-04 | Click! |
| chr13_28949369_28949858 | Sox4 | 4100 | 0.251058 | 0.45 | 3.1e-04 | Click! |
Activity of the Sox4 motif across conditions
Conditions sorted by the z-value of the Sox4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
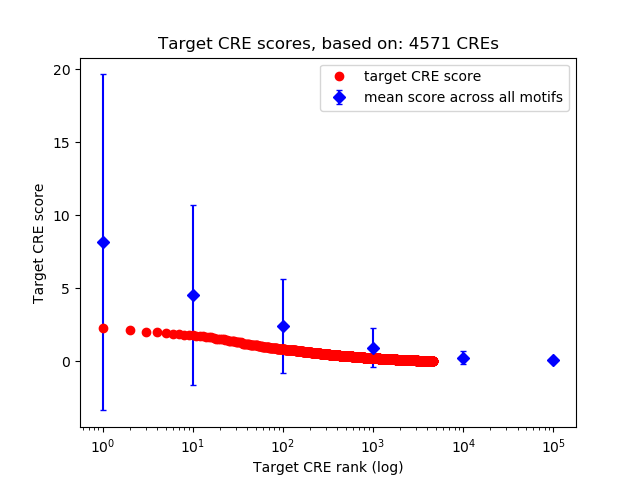
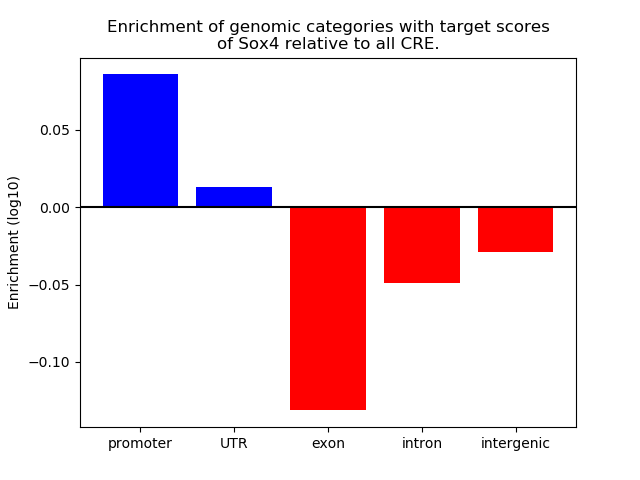
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_56634052_56634463 | 2.29 |
Lingo1 |
leucine rich repeat and Ig domain containing 1 |
1371 |
0.44 |
| chr16_43504197_43504424 | 2.14 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
613 |
0.8 |
| chr2_181764652_181765649 | 2.02 |
Myt1 |
myelin transcription factor 1 |
1818 |
0.27 |
| chrX_58032839_58033176 | 2.00 |
Zic3 |
zinc finger protein of the cerebellum 3 |
1997 |
0.41 |
| chr4_9269280_9270516 | 1.95 |
Clvs1 |
clavesin 1 |
551 |
0.81 |
| chr12_31711839_31712627 | 1.90 |
Gpr22 |
G protein-coupled receptor 22 |
1693 |
0.32 |
| chr12_52700044_52701597 | 1.87 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
1437 |
0.46 |
| chr5_65130636_65132102 | 1.83 |
Klhl5 |
kelch-like 5 |
34 |
0.97 |
| chr8_106336310_106337932 | 1.82 |
Smpd3 |
sphingomyelin phosphodiesterase 3, neutral |
867 |
0.61 |
| chr9_43070574_43070912 | 1.80 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
28111 |
0.16 |
| chr15_66239150_66239501 | 1.76 |
Kcnq3 |
potassium voltage-gated channel, subfamily Q, member 3 |
46726 |
0.14 |
| chr12_3370750_3371679 | 1.75 |
Gm48511 |
predicted gene, 48511 |
2785 |
0.18 |
| chr8_54954519_54955779 | 1.73 |
Gpm6a |
glycoprotein m6a |
306 |
0.88 |
| chr13_34126566_34127191 | 1.66 |
Tubb2b |
tubulin, beta 2B class IIB |
3476 |
0.13 |
| chr14_64589664_64590503 | 1.65 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
388 |
0.55 |
| chr17_91086370_91086959 | 1.63 |
Gm47307 |
predicted gene, 47307 |
1742 |
0.26 |
| chr2_65566848_65567533 | 1.59 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
302 |
0.92 |
| chr15_25761251_25761829 | 1.55 |
Myo10 |
myosin X |
2755 |
0.32 |
| chr15_84445152_84445303 | 1.55 |
Shisal1 |
shisa like 1 |
1318 |
0.51 |
| chr1_66176824_66177184 | 1.54 |
Map2 |
microtubule-associated protein 2 |
1454 |
0.52 |
| chr11_87759834_87761999 | 1.53 |
Tspoap1 |
TSPO associated protein 1 |
329 |
0.75 |
| chr3_88530048_88530199 | 1.51 |
Mex3a |
mex3 RNA binding family member A |
2272 |
0.12 |
| chr6_47245031_47245738 | 1.49 |
Cntnap2 |
contactin associated protein-like 2 |
931 |
0.71 |
| chr18_77560165_77560357 | 1.45 |
Rnf165 |
ring finger protein 165 |
4348 |
0.26 |
| chr13_20473087_20474265 | 1.41 |
Gm32036 |
predicted gene, 32036 |
186 |
0.89 |
| chr11_84526864_84527823 | 1.38 |
Lhx1os |
LIM homeobox 1, opposite strand |
1659 |
0.31 |
| chr15_73021529_73022598 | 1.37 |
Trappc9 |
trafficking protein particle complex 9 |
33747 |
0.18 |
| chr12_98577628_98578516 | 1.36 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
238 |
0.9 |
| chr6_137252157_137253458 | 1.36 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
342 |
0.92 |
| chr14_108910678_108911967 | 1.34 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
2836 |
0.42 |
| chr15_98169300_98169725 | 1.31 |
Ccdc184 |
coiled-coil domain containing 184 |
2354 |
0.18 |
| chr4_150651111_150652374 | 1.30 |
Slc45a1 |
solute carrier family 45, member 1 |
355 |
0.88 |
| chr16_77787990_77788627 | 1.30 |
Gm17333 |
predicted gene, 17333 |
58296 |
0.11 |
| chr8_77292695_77292846 | 1.30 |
Gm45406 |
predicted gene 45406 |
9909 |
0.18 |
| chr1_66323360_66324079 | 1.22 |
Map2 |
microtubule-associated protein 2 |
1617 |
0.37 |
| chr11_54305749_54306148 | 1.21 |
Acsl6 |
acyl-CoA synthetase long-chain family member 6 |
1746 |
0.28 |
| chr1_6760592_6761327 | 1.20 |
St18 |
suppression of tumorigenicity 18 |
23384 |
0.23 |
| chr2_178141581_178143125 | 1.19 |
Phactr3 |
phosphatase and actin regulator 3 |
420 |
0.88 |
| chrX_166347339_166348040 | 1.17 |
Gpm6b |
glycoprotein m6b |
2847 |
0.32 |
| chr10_28492781_28494073 | 1.17 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
66724 |
0.13 |
| chr2_96319240_96319943 | 1.17 |
Lrrc4c |
leucine rich repeat containing 4C |
1375 |
0.61 |
| chr13_51570228_51570409 | 1.16 |
Shc3 |
src homology 2 domain-containing transforming protein C3 |
831 |
0.67 |
| chr19_53015000_53015363 | 1.14 |
Xpnpep1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
159 |
0.95 |
| chr10_17478814_17479250 | 1.13 |
Gm47768 |
predicted gene, 47768 |
35698 |
0.16 |
| chr14_64588312_64589438 | 1.11 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
341 |
0.81 |
| chr3_8663653_8664237 | 1.11 |
Hey1 |
hairy/enhancer-of-split related with YRPW motif 1 |
2680 |
0.2 |
| chr14_103648568_103649199 | 1.10 |
Slain1 |
SLAIN motif family, member 1 |
1345 |
0.38 |
| chr2_35662845_35663372 | 1.10 |
Dab2ip |
disabled 2 interacting protein |
1489 |
0.47 |
| chr13_83725791_83726324 | 1.10 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2049 |
0.22 |
| chr12_11880981_11881577 | 1.08 |
Tubb2a-ps2 |
tubulin, beta 2a, pseudogene 2 |
1384 |
0.49 |
| chr8_64690831_64691226 | 1.08 |
Cpe |
carboxypeptidase E |
2026 |
0.3 |
| chr9_75681964_75682559 | 1.08 |
Scg3 |
secretogranin III |
1326 |
0.37 |
| chr5_124184090_124186568 | 1.06 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
211 |
0.9 |
| chr19_46326421_46327493 | 1.05 |
Psd |
pleckstrin and Sec7 domain containing |
164 |
0.86 |
| chr4_121230932_121231762 | 1.04 |
Gm12890 |
predicted gene 12890 |
3874 |
0.2 |
| chr12_12708883_12709245 | 1.04 |
Gm25538 |
predicted gene, 25538 |
9025 |
0.16 |
| chr8_15012713_15013193 | 1.02 |
Kbtbd11 |
kelch repeat and BTB (POZ) domain containing 11 |
1647 |
0.23 |
| chr3_63896453_63896604 | 1.02 |
Plch1 |
phospholipase C, eta 1 |
2845 |
0.2 |
| chr17_35568998_35569953 | 1.01 |
2300002M23Rik |
RIKEN cDNA 2300002M23 gene |
1990 |
0.14 |
| chr2_97470615_97471412 | 1.00 |
Lrrc4c |
leucine rich repeat containing 4C |
2924 |
0.4 |
| chr2_78871757_78872966 | 0.98 |
Ube2e3 |
ubiquitin-conjugating enzyme E2E 3 |
2683 |
0.32 |
| chr9_106147912_106149765 | 0.98 |
D030055H07Rik |
RIKEN cDNA D030055H07 gene |
151 |
0.9 |
| chr3_97887925_97889043 | 0.98 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
223 |
0.92 |
| chr4_5724213_5725550 | 0.97 |
Fam110b |
family with sequence similarity 110, member B |
569 |
0.81 |
| chr14_76308247_76308785 | 0.97 |
2900040C04Rik |
RIKEN cDNA 2900040C04 gene |
57059 |
0.13 |
| chr12_56276126_56276277 | 0.96 |
Gm47682 |
predicted gene, 47682 |
21803 |
0.14 |
| chr14_53173125_53173404 | 0.96 |
Gm6755 |
predicted gene 6755 |
326 |
0.85 |
| chr7_79503072_79503475 | 0.95 |
Mir9-3 |
microRNA 9-3 |
1991 |
0.16 |
| chr19_32102301_32103300 | 0.94 |
Asah2 |
N-acylsphingosine amidohydrolase 2 |
324 |
0.92 |
| chr10_109008310_109009456 | 0.94 |
Syt1 |
synaptotagmin I |
217 |
0.96 |
| chr14_52862171_52862421 | 0.94 |
Gm6740 |
predicted gene 6740 |
334 |
0.86 |
| chr3_17782537_17782894 | 0.94 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
7206 |
0.19 |
| chr9_52148115_52149635 | 0.93 |
Zc3h12c |
zinc finger CCCH type containing 12C |
19236 |
0.18 |
| chr12_69907745_69908880 | 0.92 |
Atl1 |
atlastin GTPase 1 |
580 |
0.65 |
| chr7_62277682_62277833 | 0.92 |
Gm9801 |
predicted gene 9801 |
68278 |
0.09 |
| chr16_43505394_43505961 | 0.92 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1980 |
0.41 |
| chr3_108409886_108410070 | 0.91 |
Celsr2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
5574 |
0.1 |
| chr3_8509570_8509824 | 0.90 |
Stmn2 |
stathmin-like 2 |
111 |
0.97 |
| chr15_98968974_98969326 | 0.90 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
14198 |
0.07 |
| chr4_45818603_45819141 | 0.90 |
Igfbpl1 |
insulin-like growth factor binding protein-like 1 |
2509 |
0.22 |
| chr1_66321652_66322390 | 0.90 |
Map2 |
microtubule-associated protein 2 |
23 |
0.98 |
| chr6_28827182_28827949 | 0.89 |
Lrrc4 |
leucine rich repeat containing 4 |
2780 |
0.27 |
| chr5_32153164_32153923 | 0.89 |
Fosl2 |
fos-like antigen 2 |
7022 |
0.15 |
| chr3_88213113_88214199 | 0.88 |
Gm3764 |
predicted gene 3764 |
829 |
0.3 |
| chr3_108410436_108412210 | 0.88 |
Celsr2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
4229 |
0.11 |
| chr17_90452047_90452824 | 0.88 |
Nrxn1 |
neurexin I |
2387 |
0.31 |
| chr2_158606690_158608449 | 0.87 |
Gm14204 |
predicted gene 14204 |
3021 |
0.15 |
| chr7_91262074_91263006 | 0.87 |
Gm24552 |
predicted gene, 24552 |
19545 |
0.18 |
| chr5_90902794_90902945 | 0.87 |
Cxcl2 |
chemokine (C-X-C motif) ligand 2 |
1002 |
0.41 |
| chr13_44946654_44947258 | 0.87 |
Dtnbp1 |
dystrobrevin binding protein 1 |
188 |
0.96 |
| chr14_117930688_117931113 | 0.86 |
Mir6239 |
microRNA 6239 |
22947 |
0.23 |
| chr6_112809595_112810064 | 0.85 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
19630 |
0.21 |
| chr5_107498769_107499247 | 0.85 |
Btbd8 |
BTB (POZ) domain containing 8 |
1229 |
0.33 |
| chr4_25796428_25796600 | 0.85 |
Fut9 |
fucosyltransferase 9 |
3341 |
0.24 |
| chr8_94994139_94995207 | 0.85 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
77 |
0.95 |
| chr6_21217917_21219303 | 0.84 |
Kcnd2 |
potassium voltage-gated channel, Shal-related family, member 2 |
3107 |
0.33 |
| chr11_81968072_81969328 | 0.84 |
Asic2 |
acid-sensing (proton-gated) ion channel 2 |
243 |
0.91 |
| chr5_134101487_134102203 | 0.84 |
Castor2 |
cytosolic arginine sensor for mTORC1 subunit 2 |
1840 |
0.26 |
| chr9_50725875_50726692 | 0.83 |
Dixdc1 |
DIX domain containing 1 |
1655 |
0.24 |
| chr6_12747779_12748223 | 0.83 |
Thsd7a |
thrombospondin, type I, domain containing 7A |
1252 |
0.51 |
| chr19_46506917_46507695 | 0.83 |
Trim8 |
tripartite motif-containing 8 |
5604 |
0.16 |
| chr13_42709652_42710400 | 0.83 |
Phactr1 |
phosphatase and actin regulator 1 |
445 |
0.88 |
| chr13_83732205_83734272 | 0.82 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
672 |
0.58 |
| chr6_116107038_116107576 | 0.82 |
Gm20404 |
predicted gene 20404 |
343 |
0.63 |
| chr15_98983254_98984205 | 0.81 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
102 |
0.93 |
| chr5_38683293_38683444 | 0.81 |
Zfp518b |
zinc finger protein 518B |
1382 |
0.44 |
| chr4_42953037_42954334 | 0.81 |
Dnajb5 |
DnaJ heat shock protein family (Hsp40) member B5 |
591 |
0.62 |
| chr18_16037762_16038677 | 0.80 |
Gm4835 |
predicted pseudogene 4835 |
49851 |
0.17 |
| chr15_28021392_28022419 | 0.80 |
Trio |
triple functional domain (PTPRF interacting) |
3049 |
0.28 |
| chr19_47019038_47019748 | 0.80 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
4240 |
0.14 |
| chr6_91806416_91806643 | 0.80 |
Grip2 |
glutamate receptor interacting protein 2 |
789 |
0.62 |
| chr3_107040280_107040795 | 0.79 |
AI504432 |
expressed sequence AI504432 |
1033 |
0.48 |
| chr17_35696852_35697215 | 0.79 |
Ddr1 |
discoidin domain receptor family, member 1 |
938 |
0.31 |
| chr13_95869368_95869689 | 0.79 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
22229 |
0.18 |
| chr7_62420525_62420893 | 0.79 |
Mkrn3 |
makorin, ring finger protein, 3 |
570 |
0.69 |
| chr4_32860251_32860770 | 0.79 |
Ankrd6 |
ankyrin repeat domain 6 |
68 |
0.97 |
| chr19_42409004_42409781 | 0.79 |
Gm34299 |
predicted gene, 34299 |
1002 |
0.53 |
| chr14_49524944_49526079 | 0.78 |
Slc35f4 |
solute carrier family 35, member F4 |
342 |
0.88 |
| chrX_13344663_13345577 | 0.78 |
Gm7129 |
predicted gene 7129 |
18101 |
0.15 |
| chr3_34666040_34666470 | 0.78 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
1012 |
0.36 |
| chr15_11565567_11566096 | 0.77 |
Gm49107 |
predicted gene, 49107 |
64898 |
0.12 |
| chr8_8258284_8258765 | 0.77 |
A630009H07Rik |
RIKEN cDNA A630009H07 gene |
76553 |
0.09 |
| chr5_112575572_112575844 | 0.77 |
Sez6l |
seizure related 6 homolog like |
1160 |
0.42 |
| chr12_29534253_29535510 | 0.77 |
Gm20208 |
predicted gene, 20208 |
10 |
0.8 |
| chr1_12577058_12577720 | 0.77 |
Gm2383 |
predicted gene 2383 |
11812 |
0.23 |
| chr2_73775928_73776225 | 0.76 |
Chn1 |
chimerin 1 |
730 |
0.7 |
| chr8_89036575_89038609 | 0.76 |
Sall1 |
spalt like transcription factor 1 |
6570 |
0.23 |
| chr3_51544505_51544707 | 0.76 |
Setd7 |
SET domain containing (lysine methyltransferase) 7 |
6714 |
0.11 |
| chr13_58810451_58810602 | 0.76 |
C630044B11Rik |
RIKEN cDNA C630044B11 gene |
1627 |
0.24 |
| chr1_154723254_154724531 | 0.76 |
Cacna1e |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
2028 |
0.48 |
| chr1_81076307_81076458 | 0.75 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
568 |
0.85 |
| chr19_50676799_50677457 | 0.75 |
Gm26629 |
predicted gene, 26629 |
278 |
0.88 |
| chr12_54710450_54711165 | 0.75 |
Gm22634 |
predicted gene, 22634 |
499 |
0.52 |
| chr4_30312853_30313130 | 0.75 |
Gm11924 |
predicted gene 11924 |
119121 |
0.06 |
| chr4_107679769_107680930 | 0.75 |
Dmrtb1 |
DMRT-like family B with proline-rich C-terminal, 1 |
611 |
0.63 |
| chrX_59566046_59566497 | 0.75 |
Fgf13 |
fibroblast growth factor 13 |
1201 |
0.64 |
| chr8_124795343_124796098 | 0.75 |
Trim67 |
tripartite motif-containing 67 |
2628 |
0.18 |
| chr18_43686487_43688415 | 0.74 |
Jakmip2 |
janus kinase and microtubule interacting protein 2 |
174 |
0.96 |
| chr11_105364219_105365174 | 0.74 |
Gm11638 |
predicted gene 11638 |
1444 |
0.39 |
| chr9_91352783_91353195 | 0.74 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
2036 |
0.19 |
| chr5_135725023_135726457 | 0.73 |
Por |
P450 (cytochrome) oxidoreductase |
12 |
0.96 |
| chr5_103340844_103340995 | 0.73 |
Gm42619 |
predicted gene 42619 |
3053 |
0.2 |
| chr2_149830330_149831719 | 0.73 |
Syndig1 |
synapse differentiation inducing 1 |
102 |
0.63 |
| chr4_21684238_21685103 | 0.73 |
Prdm13 |
PR domain containing 13 |
1111 |
0.49 |
| chr6_39446177_39446516 | 0.72 |
Gm25402 |
predicted gene, 25402 |
23927 |
0.11 |
| chr18_72348735_72348916 | 0.72 |
Dcc |
deleted in colorectal carcinoma |
2192 |
0.45 |
| chr5_116590520_116593206 | 0.72 |
Srrm4 |
serine/arginine repetitive matrix 4 |
46 |
0.98 |
| chr11_49168954_49169714 | 0.72 |
Btnl9 |
butyrophilin-like 9 |
1341 |
0.28 |
| chr19_36534168_36535379 | 0.72 |
Hectd2 |
HECT domain E3 ubiquitin protein ligase 2 |
19866 |
0.2 |
| chr6_8935959_8936205 | 0.72 |
Nxph1 |
neurexophilin 1 |
12349 |
0.3 |
| chr5_138275220_138276850 | 0.72 |
Gpc2 |
glypican 2 (cerebroglycan) |
2202 |
0.11 |
| chr8_73355014_73355165 | 0.71 |
Large1 |
LARGE xylosyl- and glucuronyltransferase 1 |
1549 |
0.53 |
| chr18_35855289_35856347 | 0.71 |
Cxxc5 |
CXXC finger 5 |
1080 |
0.34 |
| chr3_86844929_86845442 | 0.70 |
Gm37025 |
predicted gene, 37025 |
19307 |
0.17 |
| chr5_100120519_100121423 | 0.70 |
Tmem150c |
transmembrane protein 150C |
1825 |
0.31 |
| chr8_94869021_94869682 | 0.69 |
Dok4 |
docking protein 4 |
1853 |
0.2 |
| chr13_109928876_109929178 | 0.69 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
2183 |
0.4 |
| chr14_93888035_93888426 | 0.69 |
Pcdh9 |
protocadherin 9 |
502 |
0.87 |
| chr19_47151533_47152365 | 0.69 |
Calhm3 |
calcium homeostasis modulator 3 |
5815 |
0.1 |
| chr2_116052028_116053362 | 0.68 |
Meis2 |
Meis homeobox 2 |
3015 |
0.24 |
| chr15_98989928_98991865 | 0.68 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
5002 |
0.1 |
| chrX_10486738_10487122 | 0.68 |
Tspan7 |
tetraspanin 7 |
1772 |
0.4 |
| chr2_147181520_147181671 | 0.68 |
Nkx2-2os |
NK2 homeobox 2, opposite strand |
2488 |
0.22 |
| chr2_146324967_146325205 | 0.68 |
Gm14117 |
predicted gene 14117 |
19539 |
0.21 |
| chr18_63496060_63496613 | 0.68 |
Gm23581 |
predicted gene, 23581 |
11833 |
0.16 |
| chr3_73053153_73054210 | 0.68 |
Slitrk3 |
SLIT and NTRK-like family, member 3 |
3262 |
0.23 |
| chr13_105444000_105445296 | 0.67 |
Htr1a |
5-hydroxytryptamine (serotonin) receptor 1A |
1009 |
0.69 |
| chr3_17790851_17791260 | 0.67 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
1098 |
0.49 |
| chr12_46814495_46815083 | 0.67 |
Nova1 |
NOVA alternative splicing regulator 1 |
2171 |
0.31 |
| chr6_127336376_127337060 | 0.67 |
Gm42458 |
predicted gene 42458 |
10861 |
0.13 |
| chr6_17749231_17750103 | 0.66 |
St7 |
suppression of tumorigenicity 7 |
275 |
0.78 |
| chr4_117637193_117637344 | 0.66 |
Eri3 |
exoribonuclease 3 |
70 |
0.97 |
| chr3_144556776_144557362 | 0.66 |
Hs2st1 |
heparan sulfate 2-O-sulfotransferase 1 |
12812 |
0.14 |
| chr2_180889143_180889758 | 0.66 |
Gm14342 |
predicted gene 14342 |
210 |
0.87 |
| chr19_22447648_22448999 | 0.66 |
Trpm3 |
transient receptor potential cation channel, subfamily M, member 3 |
91 |
0.63 |
| chr6_107534114_107534926 | 0.66 |
Lrrn1 |
leucine rich repeat protein 1, neuronal |
4752 |
0.26 |
| chr16_43603833_43604204 | 0.65 |
Mir568 |
microRNA 568 |
36637 |
0.15 |
| chr1_84961778_84962209 | 0.65 |
AC167036.1 |
novel protein |
3011 |
0.17 |
| chr13_46008809_46009663 | 0.65 |
5033430I15Rik |
RIKEN cDNA 5033430I15 gene |
43885 |
0.13 |
| chr15_78116860_78117881 | 0.65 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
2336 |
0.22 |
| chr2_52511844_52512426 | 0.65 |
Gm13541 |
predicted gene 13541 |
39834 |
0.14 |
| chr3_88216674_88217001 | 0.65 |
Gm25641 |
predicted gene, 25641 |
1141 |
0.2 |
| chr4_154506617_154507505 | 0.65 |
Prdm16 |
PR domain containing 16 |
21704 |
0.18 |
| chr1_165934491_165935029 | 0.65 |
Pou2f1 |
POU domain, class 2, transcription factor 1 |
140 |
0.95 |
| chr4_116405618_116406369 | 0.64 |
Mast2 |
microtubule associated serine/threonine kinase 2 |
7 |
0.98 |
| chr5_120710211_120711265 | 0.64 |
Dtx1 |
deltex 1, E3 ubiquitin ligase |
1189 |
0.31 |
| chrX_166344270_166344507 | 0.64 |
Gpm6b |
glycoprotein m6b |
304 |
0.92 |
| chr10_119692249_119692864 | 0.63 |
Grip1 |
glutamate receptor interacting protein 1 |
502 |
0.82 |
| chr7_4836527_4837011 | 0.63 |
Shisa7 |
shisa family member 7 |
595 |
0.52 |
| chr10_14759598_14760682 | 0.63 |
Nmbr |
neuromedin B receptor |
17 |
0.91 |
| chr17_10313548_10313699 | 0.63 |
Qk |
quaking |
5738 |
0.25 |
| chr12_29528407_29529244 | 0.62 |
Myt1l |
myelin transcription factor 1-like |
424 |
0.85 |
| chr3_17793389_17793789 | 0.62 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
331 |
0.86 |
| chrX_134405492_134405850 | 0.62 |
Drp2 |
dystrophin related protein 2 |
869 |
0.59 |
| chr6_142876459_142876808 | 0.62 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
6407 |
0.21 |
| chrX_110817183_110817473 | 0.62 |
Pou3f4 |
POU domain, class 3, transcription factor 4 |
3048 |
0.3 |
| chr11_120048004_120048621 | 0.62 |
Aatk |
apoptosis-associated tyrosine kinase |
1145 |
0.3 |
| chr7_118297308_118297825 | 0.62 |
Gm23229 |
predicted gene, 23229 |
17705 |
0.13 |
| chr6_94750825_94751244 | 0.62 |
Gm43997 |
predicted gene, 43997 |
48398 |
0.11 |
| chr18_15061824_15062413 | 0.62 |
Kctd1 |
potassium channel tetramerisation domain containing 1 |
229 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 1.4 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.4 | 1.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 1.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 0.8 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.3 | 0.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 0.6 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.2 | 0.8 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 1.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.2 | 0.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 1.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 0.7 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.2 | 0.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 1.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.2 | 1.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.2 | 0.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.5 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.2 | 0.8 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 | 0.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.9 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.2 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.8 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.4 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.5 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.4 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.4 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.2 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.4 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.1 | 0.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.3 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.3 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.5 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.1 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 0.3 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 0.3 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.6 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.2 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.3 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 1.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.5 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.1 | 0.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.1 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.1 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.1 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.2 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.1 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.1 | 0.3 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.3 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.2 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 2.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.9 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.3 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.5 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 1.9 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.1 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 0.0 | 0.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0098597 | observational learning(GO:0098597) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.2 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0097476 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.6 | GO:2001258 | negative regulation of cation channel activity(GO:2001258) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 1.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.0 | GO:0097104 | postsynaptic membrane assembly(GO:0097104) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.0 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.4 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.1 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0086015 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.1 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071888 | macrophage apoptotic process(GO:0071888) regulation of macrophage apoptotic process(GO:2000109) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.0 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.0 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0071609 | chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 0.0 | 0.1 | GO:0000237 | leptotene(GO:0000237) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:2000617 | positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 0.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0072298 | regulation of metanephric glomerulus development(GO:0072298) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.0 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:1903671 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0060913 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.4 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0072679 | thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.3 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.4 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.0 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.0 | 0.0 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0031280 | negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0003284 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 0.5 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.2 | 2.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 3.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 4.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 4.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 3.1 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 1.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 0.7 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.2 | 1.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 0.8 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.2 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.1 | 0.6 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.2 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.3 | GO:0004673 | protein histidine kinase activity(GO:0004673) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.7 | GO:0043765 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.1 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.1 | 1.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 3.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |