Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
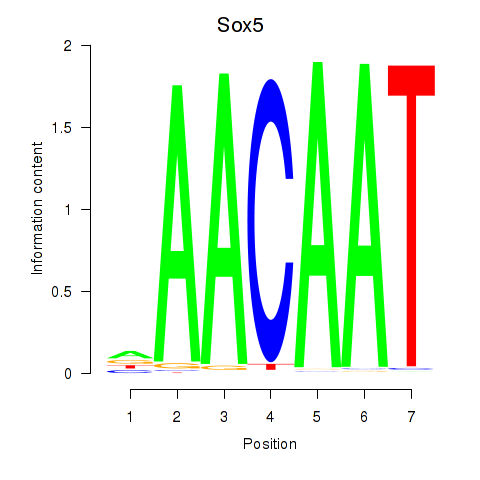
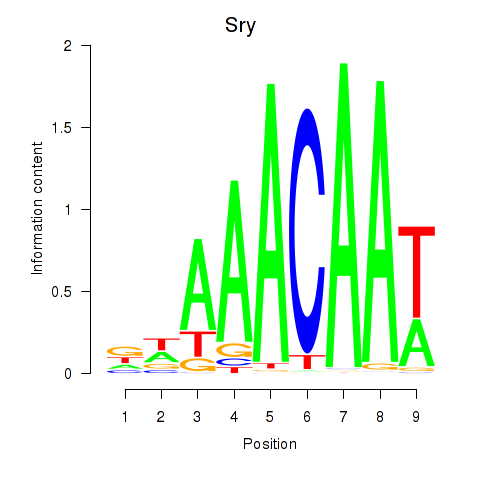
Results for Sox5_Sry
Z-value: 0.25
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSMUSG00000041540.10 | SRY (sex determining region Y)-box 5 |
|
Sry
|
ENSMUSG00000069036.3 | sex determining region of Chr Y |
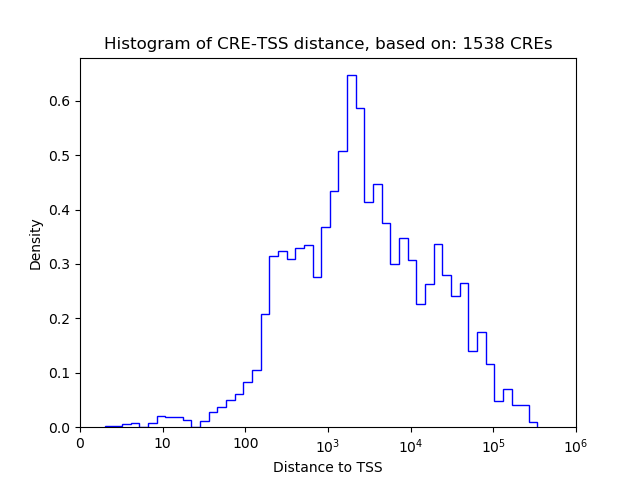
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_144180728_144181748 | Sox5 | 22819 | 0.269213 | 0.65 | 2.2e-08 | Click! |
| chr6_143832506_143833713 | Sox5 | 113979 | 0.064402 | -0.59 | 6.2e-07 | Click! |
| chr6_144782118_144782709 | Sox5 | 436 | 0.833242 | 0.37 | 3.6e-03 | Click! |
| chr6_144115538_144115689 | Sox5 | 88444 | 0.089482 | -0.36 | 4.2e-03 | Click! |
| chr6_144264338_144264733 | Sox5 | 54967 | 0.168418 | 0.36 | 4.3e-03 | Click! |
Activity of the Sox5_Sry motif across conditions
Conditions sorted by the z-value of the Sox5_Sry motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
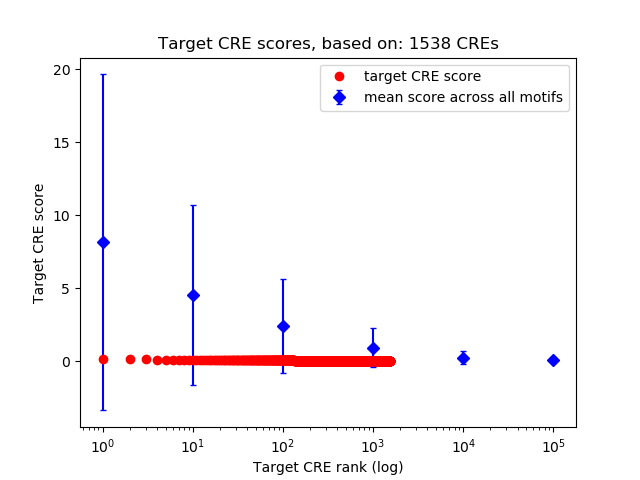
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_120022870_120023448 | 0.16 |
Xirp1 |
xin actin-binding repeat containing 1 |
439 |
0.69 |
| chr2_57698269_57698925 | 0.12 |
Gm25388 |
predicted gene, 25388 |
44775 |
0.15 |
| chr8_102783050_102783452 | 0.12 |
Gm45258 |
predicted gene 45258 |
2209 |
0.27 |
| chr13_115100230_115100381 | 0.12 |
Gm49395 |
predicted gene, 49395 |
1319 |
0.28 |
| chr13_115100442_115100733 | 0.11 |
Gm49395 |
predicted gene, 49395 |
1037 |
0.33 |
| chr7_68579903_68580131 | 0.11 |
Gm44887 |
predicted gene 44887 |
18837 |
0.2 |
| chr3_52272538_52272708 | 0.11 |
Gm20402 |
predicted gene 20402 |
3188 |
0.17 |
| chr10_13322562_13322713 | 0.11 |
Phactr2 |
phosphatase and actin regulator 2 |
1652 |
0.48 |
| chr9_32697004_32697902 | 0.10 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
1069 |
0.51 |
| chrX_96456094_96457321 | 0.10 |
Heph |
hephaestin |
279 |
0.62 |
| chr5_57723598_57724600 | 0.10 |
Gm42635 |
predicted gene 42635 |
294 |
0.83 |
| chr10_24594191_24594469 | 0.10 |
Gm15270 |
predicted gene 15270 |
572 |
0.62 |
| chr10_34485029_34485207 | 0.10 |
Frk |
fyn-related kinase |
1586 |
0.46 |
| chr14_63945494_63946032 | 0.09 |
Sox7 |
SRY (sex determining region Y)-box 7 |
2090 |
0.29 |
| chr10_125725497_125725755 | 0.09 |
Gm9102 |
predicted gene 9102 |
213095 |
0.02 |
| chr18_10786308_10786708 | 0.09 |
Mir133a-1hg |
Mir133a-1, Mir1b and Mir1a-2 host gene |
625 |
0.41 |
| chr17_48430197_48430460 | 0.09 |
Apobec2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
2290 |
0.18 |
| chr2_84422956_84423235 | 0.09 |
Calcrl |
calcitonin receptor-like |
2171 |
0.3 |
| chr3_54757529_54757680 | 0.09 |
Smad9 |
SMAD family member 9 |
2022 |
0.25 |
| chr11_116108498_116109194 | 0.09 |
Trim47 |
tripartite motif-containing 47 |
727 |
0.48 |
| chr6_29436481_29437108 | 0.09 |
Flnc |
filamin C, gamma |
3518 |
0.13 |
| chr13_40729030_40729381 | 0.09 |
Tfap2a |
transcription factor AP-2, alpha |
198 |
0.87 |
| chr1_81593421_81594539 | 0.09 |
Gm6198 |
predicted gene 6198 |
36497 |
0.2 |
| chr19_36734732_36735279 | 0.09 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1648 |
0.41 |
| chr18_40257309_40257704 | 0.09 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
483 |
0.67 |
| chr9_35424199_35424417 | 0.09 |
Cdon |
cell adhesion molecule-related/down-regulated by oncogenes |
720 |
0.64 |
| chr3_123165344_123165495 | 0.09 |
Gm43287 |
predicted gene 43287 |
947 |
0.49 |
| chr15_53342031_53342311 | 0.09 |
Ext1 |
exostosin glycosyltransferase 1 |
3488 |
0.33 |
| chr3_138198375_138198616 | 0.08 |
4930579F01Rik |
RIKEN cDNA 4930579F01 gene |
11782 |
0.13 |
| chr13_73261804_73262144 | 0.08 |
Irx4 |
Iroquois homeobox 4 |
1477 |
0.36 |
| chr7_37213357_37213873 | 0.08 |
Gm28075 |
predicted gene 28075 |
73074 |
0.09 |
| chr9_67045040_67045992 | 0.08 |
Tpm1 |
tropomyosin 1, alpha |
1554 |
0.38 |
| chr12_40198308_40199492 | 0.08 |
Gm17056 |
predicted gene 17056 |
244 |
0.64 |
| chr1_134801821_134802618 | 0.08 |
Gm37949 |
predicted gene, 37949 |
234 |
0.87 |
| chr8_87570056_87570373 | 0.08 |
4933402J07Rik |
RIKEN cDNA 4933402J07 gene |
6361 |
0.22 |
| chr17_63451168_63451380 | 0.08 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
437 |
0.88 |
| chr13_110397628_110399698 | 0.08 |
Plk2 |
polo like kinase 2 |
866 |
0.68 |
| chr2_14823792_14824714 | 0.08 |
Cacnb2 |
calcium channel, voltage-dependent, beta 2 subunit |
161 |
0.94 |
| chr10_27615824_27617033 | 0.08 |
Lama2 |
laminin, alpha 2 |
368 |
0.8 |
| chr8_48221738_48222471 | 0.08 |
Gm32842 |
predicted gene, 32842 |
48881 |
0.15 |
| chr14_28517363_28517567 | 0.08 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
6014 |
0.22 |
| chr1_192852982_192853436 | 0.08 |
Sertad4 |
SERTA domain containing 4 |
1462 |
0.26 |
| chr10_44454467_44455437 | 0.08 |
Prdm1 |
PR domain containing 1, with ZNF domain |
2258 |
0.31 |
| chr2_114051146_114052179 | 0.08 |
Actc1 |
actin, alpha, cardiac muscle 1 |
1225 |
0.4 |
| chr17_26841784_26842428 | 0.08 |
Nkx2-5 |
NK2 homeobox 5 |
541 |
0.65 |
| chr14_79107094_79107330 | 0.08 |
Gm30970 |
predicted gene, 30970 |
811 |
0.51 |
| chr16_91930196_91930347 | 0.08 |
D430001F17Rik |
RIKEN cDNA D430001F17 gene |
966 |
0.3 |
| chr15_102252882_102253053 | 0.08 |
Rarg |
retinoic acid receptor, gamma |
327 |
0.77 |
| chr2_127729042_127730365 | 0.08 |
Mall |
mal, T cell differentiation protein-like |
229 |
0.9 |
| chr5_139422893_139423950 | 0.08 |
Gper1 |
G protein-coupled estrogen receptor 1 |
141 |
0.93 |
| chr14_65326240_65326874 | 0.08 |
Gm48433 |
predicted gene, 48433 |
226 |
0.92 |
| chr16_43503039_43503193 | 0.07 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
498 |
0.85 |
| chr12_70828137_70828628 | 0.07 |
Frmd6 |
FERM domain containing 6 |
2693 |
0.24 |
| chr4_143300007_143300403 | 0.07 |
Pdpn |
podoplanin |
641 |
0.68 |
| chr4_136142605_136142974 | 0.07 |
Id3 |
inhibitor of DNA binding 3 |
708 |
0.58 |
| chr15_97004514_97004856 | 0.07 |
Slc38a4 |
solute carrier family 38, member 4 |
15266 |
0.26 |
| chr17_76484714_76484865 | 0.07 |
Gm50028 |
predicted gene, 50028 |
145594 |
0.05 |
| chr5_134988019_134988170 | 0.07 |
Cldn3 |
claudin 3 |
1880 |
0.15 |
| chr14_120279572_120279920 | 0.07 |
Mbnl2 |
muscleblind like splicing factor 2 |
4016 |
0.31 |
| chr1_187996852_187997015 | 0.07 |
Esrrg |
estrogen-related receptor gamma |
894 |
0.66 |
| chr3_34691662_34692725 | 0.07 |
Gm43206 |
predicted gene 43206 |
9560 |
0.13 |
| chr17_12768117_12768491 | 0.07 |
Igf2r |
insulin-like growth factor 2 receptor |
1360 |
0.31 |
| chr1_185481839_185482593 | 0.07 |
5033404E19Rik |
RIKEN cDNA 5033404E19 gene |
5078 |
0.16 |
| chr15_102259293_102259444 | 0.07 |
Rarg |
retinoic acid receptor, gamma |
1851 |
0.17 |
| chr3_8594088_8594846 | 0.07 |
Gm18822 |
predicted gene, 18822 |
22037 |
0.15 |
| chr17_87142136_87142409 | 0.07 |
Socs5 |
suppressor of cytokine signaling 5 |
34414 |
0.12 |
| chr6_146190446_146190623 | 0.07 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
6426 |
0.28 |
| chr16_56479571_56479912 | 0.07 |
Abi3bp |
ABI gene family, member 3 (NESH) binding protein |
1841 |
0.47 |
| chr2_68782317_68783265 | 0.07 |
Gm13612 |
predicted gene 13612 |
48226 |
0.11 |
| chr9_40717623_40717874 | 0.07 |
Gm16095 |
predicted gene 16095 |
2238 |
0.17 |
| chr3_38015645_38016389 | 0.07 |
Gm22899 |
predicted gene, 22899 |
20782 |
0.16 |
| chr16_45842068_45842739 | 0.07 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
1831 |
0.39 |
| chr18_43391464_43391742 | 0.07 |
Dpysl3 |
dihydropyrimidinase-like 3 |
1774 |
0.41 |
| chr10_23177086_23177436 | 0.07 |
Eya4 |
EYA transcriptional coactivator and phosphatase 4 |
8305 |
0.24 |
| chr5_3488483_3488698 | 0.07 |
Gm17590 |
predicted gene, 17590 |
7717 |
0.13 |
| chr12_73043399_73043995 | 0.07 |
Six1 |
sine oculis-related homeobox 1 |
882 |
0.62 |
| chr18_23497982_23498622 | 0.07 |
Dtna |
dystrobrevin alpha |
1467 |
0.56 |
| chrX_43427105_43427755 | 0.06 |
Tenm1 |
teneurin transmembrane protein 1 |
1573 |
0.42 |
| chr5_53564312_53564803 | 0.06 |
Rbpj |
recombination signal binding protein for immunoglobulin kappa J region |
8723 |
0.2 |
| chr1_162216561_162216955 | 0.06 |
Dnm3os |
dynamin 3, opposite strand |
865 |
0.43 |
| chr16_4755196_4755446 | 0.06 |
Hmox2 |
heme oxygenase 2 |
1524 |
0.27 |
| chr6_94183402_94184029 | 0.06 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
99310 |
0.08 |
| chr6_18547797_18548305 | 0.06 |
Cttnbp2 |
cortactin binding protein 2 |
33208 |
0.19 |
| chr7_89406807_89407562 | 0.06 |
Fzd4 |
frizzled class receptor 4 |
2829 |
0.19 |
| chr10_38970342_38970493 | 0.06 |
Lama4 |
laminin, alpha 4 |
4745 |
0.27 |
| chr10_5309229_5309911 | 0.06 |
Gm23573 |
predicted gene, 23573 |
47603 |
0.16 |
| chr2_85056145_85056296 | 0.06 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
1939 |
0.22 |
| chr12_119945587_119946924 | 0.06 |
Tmem196 |
transmembrane protein 196 |
155 |
0.95 |
| chr11_50381300_50381698 | 0.06 |
Hnrnph1 |
heterogeneous nuclear ribonucleoprotein H1 |
1448 |
0.31 |
| chr11_106714520_106715257 | 0.06 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
58 |
0.97 |
| chr8_25520662_25521310 | 0.06 |
Fgfr1 |
fibroblast growth factor receptor 1 |
1149 |
0.37 |
| chr1_140182332_140182724 | 0.06 |
Cfh |
complement component factor h |
752 |
0.73 |
| chr1_150992792_150993732 | 0.06 |
Hmcn1 |
hemicentin 1 |
173 |
0.95 |
| chr5_41556543_41557296 | 0.06 |
Rab28 |
RAB28, member RAS oncogene family |
70583 |
0.12 |
| chr16_29540875_29541725 | 0.06 |
Atp13a4 |
ATPase type 13A4 |
183 |
0.96 |
| chr19_32107415_32107757 | 0.06 |
Asah2 |
N-acylsphingosine amidohydrolase 2 |
563 |
0.82 |
| chr3_79683507_79683658 | 0.06 |
Gm26984 |
predicted gene, 26984 |
6706 |
0.16 |
| chr1_119348849_119349000 | 0.06 |
Gm29455 |
predicted gene 29455 |
19682 |
0.17 |
| chr11_25356867_25358335 | 0.06 |
4933427E13Rik |
RIKEN cDNA 4933427E13 gene |
27644 |
0.22 |
| chr5_32140229_32140654 | 0.06 |
Fosl2 |
fos-like antigen 2 |
4268 |
0.18 |
| chr18_32818058_32818628 | 0.06 |
Tslp |
thymic stromal lymphopoietin |
1483 |
0.34 |
| chr2_73351761_73352316 | 0.06 |
Gm13703 |
predicted gene 13703 |
5230 |
0.13 |
| chr10_4374632_4374850 | 0.06 |
Zbtb2 |
zinc finger and BTB domain containing 2 |
866 |
0.52 |
| chr5_43776922_43777113 | 0.06 |
Fbxl5 |
F-box and leucine-rich repeat protein 5 |
3417 |
0.14 |
| chr5_75155621_75156099 | 0.06 |
Pdgfra |
platelet derived growth factor receptor, alpha polypeptide |
39 |
0.89 |
| chr17_10316482_10317094 | 0.06 |
Qk |
quaking |
2573 |
0.33 |
| chr19_3685416_3685683 | 0.06 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
1007 |
0.4 |
| chr7_41979147_41979298 | 0.06 |
Vmn2r-ps57 |
vomeronasal 2, receptor, pseudogene 57 |
1706 |
0.36 |
| chr18_43841982_43842133 | 0.06 |
Gm41715 |
predicted gene, 41715 |
1961 |
0.3 |
| chr8_6907811_6908058 | 0.06 |
Gm44909 |
predicted gene 44909 |
76956 |
0.11 |
| chr1_61055351_61055642 | 0.06 |
Rpl17-ps1 |
ribosomal protein L17, pseudogene 1 |
20178 |
0.17 |
| chr2_167239298_167240893 | 0.06 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
494 |
0.75 |
| chr2_115512707_115513160 | 0.06 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
726 |
0.75 |
| chr3_99233643_99234868 | 0.06 |
Tbx15 |
T-box 15 |
6126 |
0.15 |
| chr1_62647919_62648262 | 0.06 |
Gm11600 |
predicted gene 11600 |
9698 |
0.18 |
| chr13_93612831_93613054 | 0.06 |
Gm15622 |
predicted gene 15622 |
12440 |
0.16 |
| chr5_3432895_3433605 | 0.06 |
Cdk6 |
cyclin-dependent kinase 6 |
37699 |
0.12 |
| chr3_30006216_30006441 | 0.06 |
Mecom |
MDS1 and EVI1 complex locus |
5111 |
0.22 |
| chr2_61494282_61494507 | 0.06 |
Gm22338 |
predicted gene, 22338 |
1628 |
0.51 |
| chr19_24251735_24252236 | 0.06 |
Fxn |
frataxin |
26186 |
0.14 |
| chr19_58859577_58860158 | 0.06 |
Hspa12a |
heat shock protein 12A |
771 |
0.65 |
| chr5_134742346_134742912 | 0.05 |
Gm30003 |
predicted gene, 30003 |
4413 |
0.17 |
| chr3_53525847_53526208 | 0.05 |
Frem2 |
Fras1 related extracellular matrix protein 2 |
3007 |
0.18 |
| chr13_38145014_38146012 | 0.05 |
Dsp |
desmoplakin |
5781 |
0.18 |
| chr6_116651389_116652299 | 0.05 |
Depp1 |
DEPP1 autophagy regulator |
1148 |
0.32 |
| chr2_33218844_33219764 | 0.05 |
Angptl2 |
angiopoietin-like 2 |
3187 |
0.21 |
| chr4_34328044_34328577 | 0.05 |
Gm12751 |
predicted gene 12751 |
25733 |
0.18 |
| chr9_24766942_24767354 | 0.05 |
Tbx20 |
T-box 20 |
2532 |
0.28 |
| chr3_137343040_137343246 | 0.05 |
Emcn |
endomucin |
1969 |
0.42 |
| chr2_76982792_76983214 | 0.05 |
Ttn |
titin |
456 |
0.86 |
| chr10_44452907_44453510 | 0.05 |
Prdm1 |
PR domain containing 1, with ZNF domain |
4002 |
0.23 |
| chr6_15405115_15405989 | 0.05 |
Gm25470 |
predicted gene, 25470 |
571 |
0.83 |
| chr15_91709273_91710162 | 0.05 |
Lrrk2 |
leucine-rich repeat kinase 2 |
29231 |
0.19 |
| chr12_95691954_95693678 | 0.05 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
590 |
0.73 |
| chr3_154269822_154270217 | 0.05 |
Slc44a5 |
solute carrier family 44, member 5 |
12545 |
0.18 |
| chr17_12763317_12763493 | 0.05 |
Igf2r |
insulin-like growth factor 2 receptor |
6259 |
0.13 |
| chr9_116173153_116173304 | 0.05 |
Tgfbr2 |
transforming growth factor, beta receptor II |
2037 |
0.31 |
| chrX_114476657_114476851 | 0.05 |
Klhl4 |
kelch-like 4 |
2150 |
0.37 |
| chr10_21924319_21924470 | 0.05 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
4076 |
0.23 |
| chr2_62572999_62574382 | 0.05 |
Fap |
fibroblast activation protein |
130 |
0.97 |
| chr6_147086429_147087322 | 0.05 |
Mansc4 |
MANSC domain containing 4 |
126 |
0.95 |
| chr13_53283065_53283604 | 0.05 |
Ror2 |
receptor tyrosine kinase-like orphan receptor 2 |
2215 |
0.39 |
| chr10_11250316_11250467 | 0.05 |
4930432B10Rik |
RIKEN cDNA 4930432B10 gene |
30607 |
0.12 |
| chr1_58930482_58930757 | 0.05 |
Trak2 |
trafficking protein, kinesin binding 2 |
8922 |
0.16 |
| chr4_32430676_32431345 | 0.05 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
13575 |
0.26 |
| chr16_20099317_20099468 | 0.05 |
Klhl24 |
kelch-like 24 |
1800 |
0.36 |
| chr1_14308374_14310407 | 0.05 |
Eya1 |
EYA transcriptional coactivator and phosphatase 1 |
446 |
0.88 |
| chr9_35423074_35423780 | 0.05 |
Cdon |
cell adhesion molecule-related/down-regulated by oncogenes |
161 |
0.95 |
| chr14_90028075_90028226 | 0.05 |
Gm48941 |
predicted gene, 48941 |
57742 |
0.15 |
| chr5_43776669_43776821 | 0.05 |
Fbxl5 |
F-box and leucine-rich repeat protein 5 |
3145 |
0.15 |
| chr1_163358065_163358620 | 0.05 |
Gm24940 |
predicted gene, 24940 |
43941 |
0.11 |
| chr10_85183402_85183765 | 0.05 |
Cry1 |
cryptochrome 1 (photolyase-like) |
1481 |
0.42 |
| chr10_103021254_103021450 | 0.05 |
Alx1 |
ALX homeobox 1 |
1222 |
0.49 |
| chr3_89688580_89688818 | 0.05 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
26181 |
0.13 |
| chr2_6873585_6874006 | 0.05 |
Celf2 |
CUGBP, Elav-like family member 2 |
1198 |
0.49 |
| chr10_53377941_53378236 | 0.05 |
Gm47641 |
predicted gene, 47641 |
1364 |
0.29 |
| chr7_27168286_27168475 | 0.05 |
Egln2 |
egl-9 family hypoxia-inducible factor 2 |
1578 |
0.16 |
| chr5_92899677_92900334 | 0.05 |
Shroom3 |
shroom family member 3 |
2012 |
0.38 |
| chr3_149844857_149845745 | 0.05 |
Gm31121 |
predicted gene, 31121 |
184905 |
0.03 |
| chr2_172934875_172936042 | 0.05 |
Bmp7 |
bone morphogenetic protein 7 |
4634 |
0.21 |
| chr5_13628238_13629170 | 0.05 |
Gm43718 |
predicted gene 43718 |
10889 |
0.19 |
| chr16_25802021_25802276 | 0.05 |
Trp63 |
transformation related protein 63 |
232 |
0.96 |
| chr10_7182673_7182942 | 0.05 |
Cnksr3 |
Cnksr family member 3 |
29430 |
0.23 |
| chr9_87724164_87724694 | 0.05 |
D030062O11Rik |
RIKEN cDNA D030062O11 gene |
4856 |
0.19 |
| chr14_61043748_61044093 | 0.05 |
Tnfrsf19 |
tumor necrosis factor receptor superfamily, member 19 |
2549 |
0.3 |
| chr13_59073511_59074258 | 0.05 |
Gm34245 |
predicted gene, 34245 |
4412 |
0.21 |
| chr1_82284367_82285219 | 0.05 |
Irs1 |
insulin receptor substrate 1 |
6623 |
0.2 |
| chr5_17642536_17642831 | 0.05 |
Sema3c |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
51519 |
0.17 |
| chr8_108241119_108241834 | 0.05 |
Zfhx3 |
zinc finger homeobox 3 |
246917 |
0.02 |
| chr17_10310558_10310848 | 0.05 |
Qk |
quaking |
8658 |
0.24 |
| chr6_118478476_118479294 | 0.05 |
Zfp9 |
zinc finger protein 9 |
435 |
0.79 |
| chr5_131524957_131525108 | 0.05 |
Auts2 |
autism susceptibility candidate 2 |
9365 |
0.2 |
| chr2_30864765_30865574 | 0.05 |
Prrx2 |
paired related homeobox 2 |
14150 |
0.12 |
| chr9_63754826_63756126 | 0.05 |
Smad3 |
SMAD family member 3 |
2518 |
0.33 |
| chr10_81363483_81364417 | 0.05 |
4930404N11Rik |
RIKEN cDNA 4930404N11 gene |
904 |
0.26 |
| chr3_50443042_50444382 | 0.05 |
Slc7a11 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
98 |
0.98 |
| chr14_64834470_64835221 | 0.05 |
Gm20111 |
predicted gene, 20111 |
4255 |
0.18 |
| chr8_46502513_46502977 | 0.05 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
9913 |
0.16 |
| chr2_70934994_70935182 | 0.05 |
Mettl8 |
methyltransferase like 8 |
38300 |
0.15 |
| chr3_144714823_144715024 | 0.05 |
Sh3glb1 |
SH3-domain GRB2-like B1 (endophilin) |
5187 |
0.15 |
| chr4_32302507_32304025 | 0.05 |
Bach2it1 |
BTB and CNC homology 2, intronic transcript 1 |
51505 |
0.14 |
| chr2_174858675_174858999 | 0.05 |
Gm14616 |
predicted gene 14616 |
2548 |
0.24 |
| chr2_44271339_44271725 | 0.05 |
Gm22867 |
predicted gene, 22867 |
157403 |
0.04 |
| chr3_147999482_147999853 | 0.05 |
Gm43574 |
predicted gene 43574 |
130577 |
0.06 |
| chr6_71629954_71630268 | 0.05 |
Kdm3a |
lysine (K)-specific demethylase 3A |
2575 |
0.2 |
| chr18_6513819_6514449 | 0.05 |
Epc1 |
enhancer of polycomb homolog 1 |
1974 |
0.29 |
| chrX_103185752_103186844 | 0.05 |
Nap1l2 |
nucleosome assembly protein 1-like 2 |
342 |
0.83 |
| chr4_88935794_88936347 | 0.05 |
Gm49890 |
predicted gene, 49890 |
2451 |
0.18 |
| chr16_42189565_42189855 | 0.05 |
Gm49737 |
predicted gene, 49737 |
4793 |
0.23 |
| chr5_139128762_139129722 | 0.05 |
Prkar1b |
protein kinase, cAMP dependent regulatory, type I beta |
517 |
0.75 |
| chr4_84671699_84671986 | 0.05 |
Bnc2 |
basonuclin 2 |
3154 |
0.3 |
| chr12_108837523_108838185 | 0.05 |
Slc25a47 |
solute carrier family 25, member 47 |
790 |
0.33 |
| chr18_47370463_47370942 | 0.05 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
1832 |
0.37 |
| chr10_8761475_8762950 | 0.05 |
Sash1 |
SAM and SH3 domain containing 1 |
262 |
0.93 |
| chr9_66806165_66806354 | 0.05 |
Ppp1r2-ps4 |
protein phosphatase 1, regulatory (inhibitor) subunit 2, pseudogene 4 |
321 |
0.8 |
| chr5_66524169_66524487 | 0.05 |
Gm43343 |
predicted gene 43343 |
1784 |
0.29 |
| chr3_127525390_127525541 | 0.05 |
Gm43354 |
predicted gene 43354 |
12565 |
0.07 |
| chr2_105616227_105616777 | 0.05 |
Paupar |
Pax6 upstream antisense RNA |
44841 |
0.11 |
| chr18_10707052_10707203 | 0.05 |
4930563E18Rik |
RIKEN cDNA 4930563E18 gene |
267 |
0.57 |
| chr19_59941623_59942210 | 0.05 |
Rab11fip2 |
RAB11 family interacting protein 2 (class I) |
1084 |
0.46 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |