Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
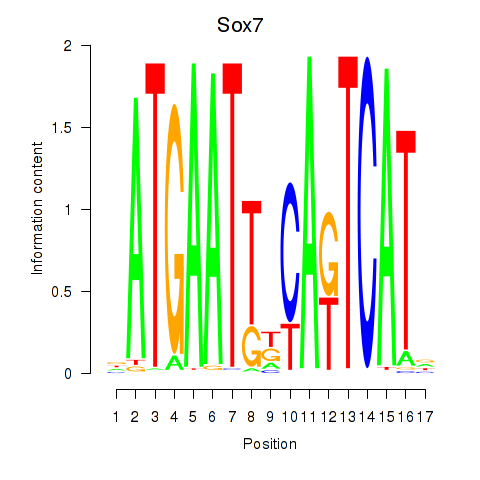
Results for Sox7
Z-value: 0.95
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSMUSG00000063060.5 | SRY (sex determining region Y)-box 7 |
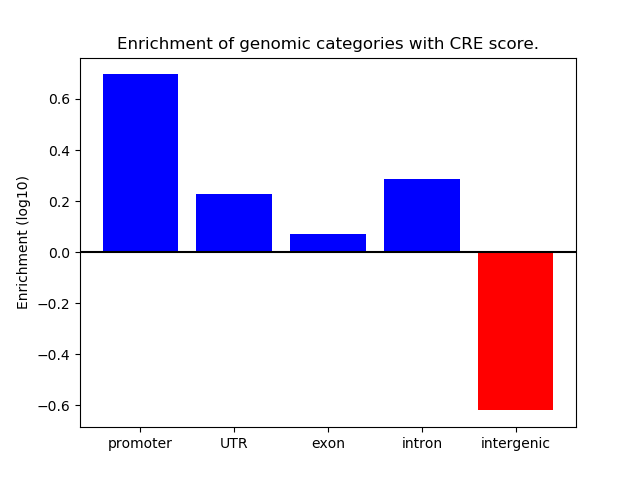
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_63941790_63942313 | Sox7 | 1622 | 0.359212 | -0.43 | 5.4e-04 | Click! |
| chr14_63940714_63941205 | Sox7 | 2714 | 0.251160 | -0.36 | 5.1e-03 | Click! |
| chr14_63942562_63942943 | Sox7 | 921 | 0.561285 | -0.31 | 1.5e-02 | Click! |
| chr14_63943010_63944396 | Sox7 | 30 | 0.975591 | -0.30 | 1.9e-02 | Click! |
| chr14_63941610_63941761 | Sox7 | 1988 | 0.307388 | -0.22 | 8.6e-02 | Click! |
Activity of the Sox7 motif across conditions
Conditions sorted by the z-value of the Sox7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
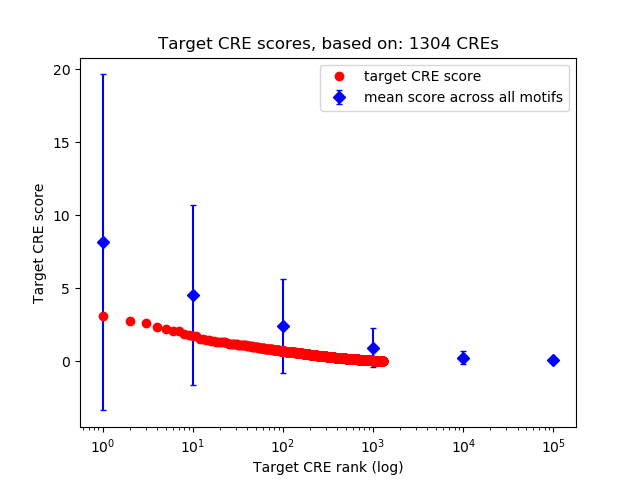
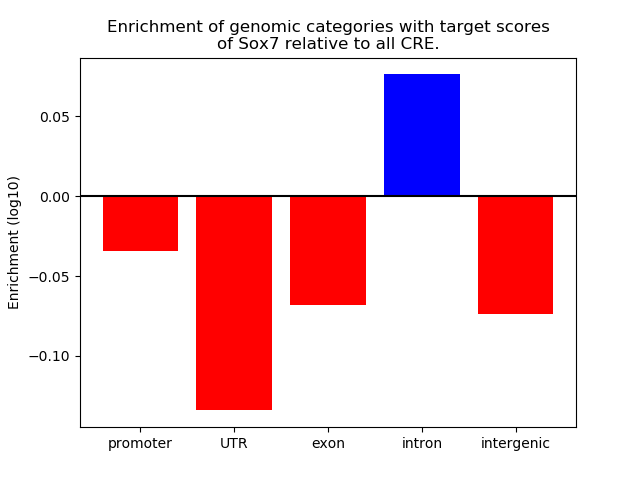
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_112232861_112233588 | 3.07 |
Arpp21 |
cyclic AMP-regulated phosphoprotein, 21 |
402 |
0.75 |
| chr6_103513736_103514218 | 2.77 |
Chl1 |
cell adhesion molecule L1-like |
2647 |
0.25 |
| chr2_121803809_121804141 | 2.65 |
Frmd5 |
FERM domain containing 5 |
2897 |
0.26 |
| chr4_22483966_22484156 | 2.32 |
Pou3f2 |
POU domain, class 3, transcription factor 2 |
4305 |
0.19 |
| chr7_142091808_142093330 | 2.23 |
Dusp8 |
dual specificity phosphatase 8 |
2703 |
0.11 |
| chr8_109245493_109246323 | 2.11 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
3958 |
0.33 |
| chr13_84063384_84064052 | 2.06 |
Gm17750 |
predicted gene, 17750 |
1054 |
0.58 |
| chr2_97472737_97472888 | 1.87 |
Lrrc4c |
leucine rich repeat containing 4C |
4723 |
0.34 |
| chr12_86680036_86680970 | 1.79 |
Vash1 |
vasohibin 1 |
1803 |
0.28 |
| chr12_26472511_26472823 | 1.76 |
Cmpk2 |
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
1263 |
0.39 |
| chr1_81077232_81078427 | 1.71 |
Nyap2 |
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
246 |
0.96 |
| chr13_83717521_83718816 | 1.53 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
3213 |
0.17 |
| chr1_6731579_6732076 | 1.49 |
St18 |
suppression of tumorigenicity 18 |
1725 |
0.48 |
| chr13_78172031_78172578 | 1.49 |
3110006O06Rik |
RIKEN cDNA 3110006O06 gene |
674 |
0.61 |
| chr7_61309776_61310205 | 1.43 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
1723 |
0.5 |
| chr4_22497544_22499061 | 1.42 |
Gm30731 |
predicted gene, 30731 |
7754 |
0.16 |
| chr12_46817370_46817521 | 1.41 |
Nova1 |
NOVA alternative splicing regulator 1 |
485 |
0.82 |
| chr13_21382104_21382258 | 1.35 |
Gm11271 |
predicted gene 11271 |
1051 |
0.29 |
| chr2_94243013_94244880 | 1.34 |
Mir670hg |
MIR670 host gene (non-protein coding) |
608 |
0.62 |
| chr11_94995250_94996098 | 1.34 |
Ppp1r9b |
protein phosphatase 1, regulatory subunit 9B |
390 |
0.75 |
| chr5_22341654_22342038 | 1.33 |
Reln |
reelin |
2844 |
0.2 |
| chr5_9722064_9722215 | 1.30 |
Grm3 |
glutamate receptor, metabotropic 3 |
3031 |
0.3 |
| chr10_39729709_39729860 | 1.29 |
E130307A14Rik |
RIKEN cDNA E130307A14 gene |
1603 |
0.27 |
| chr7_103825389_103825783 | 1.25 |
Hbb-bs |
hemoglobin, beta adult s chain |
2139 |
0.11 |
| chrX_9019650_9019801 | 1.21 |
Gm9431 |
predicted gene 9431 |
9648 |
0.07 |
| chr10_105689563_105689778 | 1.19 |
n-R5s80 |
nuclear encoded rRNA 5S 80 |
114527 |
0.05 |
| chr13_115911109_115911441 | 1.18 |
Gm18135 |
predicted gene, 18135 |
140339 |
0.05 |
| chr10_63276979_63277164 | 1.18 |
Herc4 |
hect domain and RLD 4 |
1332 |
0.31 |
| chr16_8540255_8540422 | 1.16 |
Abat |
4-aminobutyrate aminotransferase |
26853 |
0.14 |
| chr10_23674717_23675319 | 1.15 |
4930520K02Rik |
RIKEN cDNA 4930520K02 gene |
52870 |
0.11 |
| chr5_51566574_51566861 | 1.15 |
Ppargc1a |
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
987 |
0.6 |
| chr15_85676712_85677176 | 1.14 |
Lncppara |
long noncoding RNA near Ppara |
23328 |
0.12 |
| chr19_36054870_36055220 | 1.12 |
Htr7 |
5-hydroxytryptamine (serotonin) receptor 7 |
2295 |
0.33 |
| chr13_8211692_8211843 | 1.12 |
Adarb2 |
adenosine deaminase, RNA-specific, B2 |
8845 |
0.18 |
| chr15_9529382_9529634 | 1.12 |
Il7r |
interleukin 7 receptor |
258 |
0.94 |
| chr3_16817433_16817601 | 1.10 |
Gm26485 |
predicted gene, 26485 |
5795 |
0.35 |
| chr4_108299089_108299240 | 1.09 |
Zyg11b |
zyg-ll family member B, cell cycle regulator |
1932 |
0.25 |
| chr1_194622071_194623282 | 1.09 |
Plxna2 |
plexin A2 |
2851 |
0.26 |
| chr4_97788585_97788930 | 1.09 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
10679 |
0.2 |
| chr6_86079772_86080184 | 1.08 |
Add2 |
adducin 2 (beta) |
1894 |
0.25 |
| chr3_17582970_17583121 | 1.07 |
Gm38154 |
predicted gene, 38154 |
87643 |
0.09 |
| chr9_74864280_74865379 | 1.07 |
Onecut1 |
one cut domain, family member 1 |
1655 |
0.3 |
| chr11_37561013_37561164 | 1.05 |
Gm12128 |
predicted gene 12128 |
97901 |
0.09 |
| chr16_16558986_16560577 | 1.04 |
Fgd4 |
FYVE, RhoGEF and PH domain containing 4 |
209 |
0.94 |
| chr10_95790859_95791151 | 1.01 |
4732465J04Rik |
RIKEN cDNA 4732465J04 gene |
12088 |
0.12 |
| chr13_83714553_83714869 | 1.00 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
6670 |
0.14 |
| chr2_169998849_169999000 | 0.99 |
AY702102 |
cDNA sequence AY702102 |
35737 |
0.21 |
| chr5_8621588_8621754 | 0.99 |
Rundc3b |
RUN domain containing 3B |
1281 |
0.47 |
| chr9_88550647_88551163 | 0.98 |
Zfp949 |
zinc finger protein 949 |
2722 |
0.14 |
| chr10_73379238_73379395 | 0.98 |
Gm19168 |
predicted gene, 19168 |
3283 |
0.31 |
| chr9_41589121_41589444 | 0.98 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
788 |
0.48 |
| chr8_31939339_31939953 | 0.95 |
Nrg1 |
neuregulin 1 |
10389 |
0.25 |
| chr15_88290858_88291278 | 0.95 |
B230214G05Rik |
RIKEN cDNA B230214G05 gene |
23806 |
0.21 |
| chr3_17800898_17801130 | 0.91 |
Gm23441 |
predicted gene, 23441 |
2733 |
0.24 |
| chr2_131911163_131911433 | 0.91 |
Prn |
prion protein readthrough transcript |
1341 |
0.27 |
| chr13_71157717_71158937 | 0.90 |
Mir466f-4 |
microRNA 466f-4 |
51238 |
0.17 |
| chr1_137606248_137606653 | 0.90 |
Gm37903 |
predicted gene, 37903 |
75220 |
0.1 |
| chr4_46854379_46855929 | 0.89 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
4748 |
0.3 |
| chr12_40447960_40448111 | 0.89 |
Dock4 |
dedicator of cytokinesis 4 |
1699 |
0.36 |
| chr10_29531048_29531256 | 0.89 |
Gm48159 |
predicted gene, 48159 |
3565 |
0.21 |
| chr2_169445439_169445840 | 0.88 |
Gm14249 |
predicted gene 14249 |
34978 |
0.16 |
| chr16_42339015_42340584 | 0.88 |
Gap43 |
growth associated protein 43 |
852 |
0.7 |
| chr4_88029546_88030118 | 0.88 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
2981 |
0.22 |
| chr8_41054476_41055299 | 0.87 |
Mtus1 |
mitochondrial tumor suppressor 1 |
93 |
0.95 |
| chrX_95476704_95476855 | 0.86 |
Asb12 |
ankyrin repeat and SOCS box-containing 12 |
1350 |
0.47 |
| chr15_58889728_58889985 | 0.86 |
Gm49356 |
predicted gene, 49356 |
558 |
0.44 |
| chr14_99521583_99522333 | 0.86 |
Gm41230 |
predicted gene, 41230 |
587 |
0.8 |
| chr6_13832798_13833136 | 0.85 |
Gpr85 |
G protein-coupled receptor 85 |
4274 |
0.24 |
| chr5_133571082_133571461 | 0.84 |
Gm36667 |
predicted gene, 36667 |
27135 |
0.21 |
| chrX_164439505_164439752 | 0.83 |
Asb11 |
ankyrin repeat and SOCS box-containing 11 |
1570 |
0.35 |
| chr5_70841090_70841579 | 0.83 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
870 |
0.75 |
| chr4_134866605_134866756 | 0.83 |
Rhd |
Rh blood group, D antigen |
2144 |
0.27 |
| chr1_6732598_6732857 | 0.83 |
St18 |
suppression of tumorigenicity 18 |
2143 |
0.43 |
| chr13_34340668_34341142 | 0.82 |
Slc22a23 |
solute carrier family 22, member 23 |
3298 |
0.23 |
| chr5_17835023_17835212 | 0.82 |
Cd36 |
CD36 molecule |
579 |
0.85 |
| chr5_52440219_52440498 | 0.82 |
Gm3519 |
predicted gene 3519 |
801 |
0.62 |
| chr8_108535196_108536010 | 0.81 |
Gm39244 |
predicted gene, 39244 |
1344 |
0.54 |
| chr9_42460457_42460722 | 0.80 |
Tbcel |
tubulin folding cofactor E-like |
872 |
0.57 |
| chrX_144685908_144686546 | 0.79 |
Trpc5 |
transient receptor potential cation channel, subfamily C, member 5 |
1776 |
0.37 |
| chr2_105675959_105678109 | 0.79 |
Pax6 |
paired box 6 |
905 |
0.54 |
| chr1_42230776_42231106 | 0.79 |
Gm9915 |
predicted gene 9915 |
1214 |
0.46 |
| chr9_91404809_91406365 | 0.79 |
Gm29478 |
predicted gene 29478 |
1113 |
0.42 |
| chr13_78193022_78193812 | 0.78 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
2956 |
0.18 |
| chr16_4414999_4415926 | 0.78 |
Adcy9 |
adenylate cyclase 9 |
4125 |
0.25 |
| chr2_45111564_45111715 | 0.77 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
465 |
0.57 |
| chr5_66678971_66679451 | 0.77 |
Uchl1 |
ubiquitin carboxy-terminal hydrolase L1 |
2319 |
0.2 |
| chr8_12400578_12402091 | 0.77 |
Gm25239 |
predicted gene, 25239 |
4931 |
0.15 |
| chr11_46448271_46448491 | 0.76 |
Gm12168 |
predicted gene 12168 |
2966 |
0.17 |
| chr1_172028424_172029004 | 0.76 |
Vangl2 |
VANGL planar cell polarity 2 |
270 |
0.87 |
| chr11_29170569_29170729 | 0.74 |
Ppp4r3b |
protein phosphatase 4 regulatory subunit 3B |
2241 |
0.26 |
| chr10_90043418_90043569 | 0.74 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
3129 |
0.37 |
| chr5_125056018_125058841 | 0.74 |
Gm42838 |
predicted gene 42838 |
412 |
0.71 |
| chr12_29724156_29724307 | 0.73 |
C630031E19Rik |
RIKEN cDNA C630031E19 gene |
37786 |
0.21 |
| chr7_85289417_85289568 | 0.73 |
Gm20415 |
predicted gene 20415 |
119 |
0.96 |
| chr6_86636670_86636892 | 0.73 |
Asprv1 |
aspartic peptidase, retroviral-like 1 |
8617 |
0.11 |
| chr15_51363475_51363626 | 0.73 |
Gm19303 |
predicted gene, 19303 |
32060 |
0.24 |
| chr1_160060957_160061168 | 0.72 |
4930523C07Rik |
RIKEN cDNA 4930523C07 gene |
14100 |
0.15 |
| chr19_10303270_10303421 | 0.72 |
Dagla |
diacylglycerol lipase, alpha |
1512 |
0.33 |
| chr3_67892003_67892637 | 0.71 |
Iqschfp |
Iqcj and Schip1 fusion protein |
88 |
0.51 |
| chr8_93814307_93815014 | 0.71 |
4930488L21Rik |
RIKEN cDNA 4930488L21 gene |
938 |
0.54 |
| chr6_58905914_58906262 | 0.70 |
Nap1l5 |
nucleosome assembly protein 1-like 5 |
585 |
0.62 |
| chr18_35210523_35210985 | 0.69 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
4176 |
0.2 |
| chr7_24529166_24529317 | 0.69 |
AC161166.1 |
zinc finger protein 576 (ZNF576) pseudogene |
601 |
0.47 |
| chr13_96668711_96668944 | 0.69 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
827 |
0.56 |
| chr6_88803414_88803565 | 0.68 |
Gm44001 |
predicted gene, 44001 |
1246 |
0.32 |
| chr11_38681045_38681331 | 0.67 |
Gm23520 |
predicted gene, 23520 |
117089 |
0.07 |
| chr10_106472759_106472910 | 0.67 |
Ppfia2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
2488 |
0.3 |
| chr2_57697097_57697716 | 0.66 |
Gm25388 |
predicted gene, 25388 |
43584 |
0.16 |
| chr11_35977679_35978225 | 0.66 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
2575 |
0.32 |
| chr12_49397455_49398044 | 0.66 |
3110039M20Rik |
RIKEN cDNA 3110039M20 gene |
7090 |
0.14 |
| chr12_108425010_108425685 | 0.66 |
Eml1 |
echinoderm microtubule associated protein like 1 |
2269 |
0.3 |
| chr5_143548758_143550004 | 0.66 |
Fam220a |
family with sequence similarity 220, member A |
341 |
0.84 |
| chr7_61220273_61220470 | 0.66 |
Gm38451 |
predicted gene, 38451 |
89687 |
0.08 |
| chr10_93695291_93695671 | 0.65 |
Gm15915 |
predicted gene 15915 |
12159 |
0.14 |
| chr5_57723103_57723321 | 0.65 |
Pcdh7 |
protocadherin 7 |
1003 |
0.34 |
| chrX_64810691_64810842 | 0.64 |
Gm26111 |
predicted gene, 26111 |
10255 |
0.23 |
| chr3_104217848_104218047 | 0.64 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
2156 |
0.19 |
| chr2_37264254_37264405 | 0.64 |
Olfr367-ps |
olfactory receptor 367, pseudogene |
2622 |
0.2 |
| chr5_90655963_90656577 | 0.64 |
Rassf6 |
Ras association (RalGDS/AF-6) domain family member 6 |
15613 |
0.16 |
| chr7_132262164_132262500 | 0.63 |
Chst15 |
carbohydrate sulfotransferase 15 |
16293 |
0.17 |
| chr13_83727321_83728283 | 0.63 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
304 |
0.83 |
| chr1_136960004_136960405 | 0.63 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
176 |
0.96 |
| chr7_78578604_78579236 | 0.63 |
Gm9885 |
predicted gene 9885 |
90 |
0.69 |
| chrX_135887222_135887438 | 0.63 |
Bhlhb9 |
basic helix-loop-helix domain containing, class B9 |
1347 |
0.42 |
| chr9_113507383_113507910 | 0.63 |
AU023762 |
expressed sequence AU023762 |
709 |
0.71 |
| chr14_77158532_77158695 | 0.63 |
Enox1 |
ecto-NOX disulfide-thiol exchanger 1 |
1833 |
0.41 |
| chr4_9273042_9273193 | 0.62 |
Clvs1 |
clavesin 1 |
1662 |
0.42 |
| chr7_77445511_77445911 | 0.62 |
Gm44608 |
predicted gene 44608 |
33138 |
0.25 |
| chrX_60146958_60147936 | 0.61 |
Mcf2 |
mcf.2 transforming sequence |
196 |
0.96 |
| chr6_55680954_55681113 | 0.61 |
Neurod6 |
neurogenic differentiation 6 |
230 |
0.94 |
| chr14_45221914_45222158 | 0.61 |
Txndc16 |
thioredoxin domain containing 16 |
1708 |
0.23 |
| chr7_35837426_35837577 | 0.61 |
Gm28514 |
predicted gene 28514 |
759 |
0.67 |
| chr14_100026766_100027149 | 0.61 |
Klf12 |
Kruppel-like factor 12 |
83249 |
0.1 |
| chr12_8561626_8561925 | 0.61 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
12154 |
0.18 |
| chr8_12388354_12388751 | 0.60 |
Gm45560 |
predicted gene 45560 |
1616 |
0.27 |
| chr2_85431998_85432149 | 0.60 |
Olfr994 |
olfactory receptor 994 |
1246 |
0.27 |
| chr12_107999037_107999696 | 0.60 |
Bcl11b |
B cell leukemia/lymphoma 11B |
4048 |
0.33 |
| chr8_60952040_60952217 | 0.60 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
2620 |
0.24 |
| chr10_126002862_126003306 | 0.60 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
8547 |
0.26 |
| chr13_109927479_109928182 | 0.59 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
986 |
0.65 |
| chr14_36966037_36966188 | 0.59 |
Ccser2 |
coiled-coil serine rich 2 |
2575 |
0.3 |
| chr2_79451460_79451814 | 0.59 |
Neurod1 |
neurogenic differentiation 1 |
5114 |
0.23 |
| chr9_16498247_16498398 | 0.59 |
Fat3 |
FAT atypical cadherin 3 |
2963 |
0.38 |
| chr4_27981464_27981615 | 0.58 |
Tpm3-rs2 |
tropomyosin 3, related sequence 2 |
15366 |
0.27 |
| chr12_116283592_116283802 | 0.58 |
Esyt2 |
extended synaptotagmin-like protein 2 |
2446 |
0.19 |
| chr9_102720016_102720747 | 0.58 |
Amotl2 |
angiomotin-like 2 |
94 |
0.95 |
| chr15_59079090_59079241 | 0.58 |
Mtss1 |
MTSS I-BAR domain containing 1 |
2824 |
0.32 |
| chr7_54069871_54070022 | 0.58 |
Gm27921 |
predicted gene, 27921 |
61366 |
0.14 |
| chr8_23414255_23415055 | 0.57 |
Sfrp1 |
secreted frizzled-related protein 1 |
3153 |
0.33 |
| chr17_80946883_80947193 | 0.57 |
Tmem178 |
transmembrane protein 178 |
2406 |
0.31 |
| chr15_85677376_85679232 | 0.57 |
Lncppara |
long noncoding RNA near Ppara |
24688 |
0.12 |
| chrX_169829854_169830387 | 0.56 |
Mid1 |
midline 1 |
1961 |
0.41 |
| chr19_19058784_19059088 | 0.56 |
Rorb |
RAR-related orphan receptor beta |
52260 |
0.17 |
| chr6_114289017_114289311 | 0.56 |
Slc6a1 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 1 |
6374 |
0.27 |
| chr2_106716257_106716636 | 0.56 |
Mpped2 |
metallophosphoesterase domain containing 2 |
14044 |
0.23 |
| chr6_131783861_131784012 | 0.56 |
Tas2r114 |
taste receptor, type 2, member 114 |
93873 |
0.05 |
| chr4_19441647_19441884 | 0.55 |
Cpne3 |
copine III |
101913 |
0.07 |
| chr15_79684785_79686121 | 0.55 |
Josd1 |
Josephin domain containing 1 |
2466 |
0.13 |
| chr10_77220323_77220656 | 0.55 |
Pofut2 |
protein O-fucosyltransferase 2 |
38729 |
0.12 |
| chr2_135712427_135712633 | 0.54 |
Gm14211 |
predicted gene 14211 |
19376 |
0.18 |
| chr9_4793875_4794307 | 0.54 |
Gria4 |
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
1428 |
0.59 |
| chr13_100743804_100744148 | 0.54 |
Mrps36 |
mitochondrial ribosomal protein S36 |
137 |
0.93 |
| chr19_37177777_37178526 | 0.54 |
Cpeb3 |
cytoplasmic polyadenylation element binding protein 3 |
134 |
0.91 |
| chr2_120409901_120410052 | 0.54 |
Ganc |
glucosidase, alpha; neutral C |
1437 |
0.34 |
| chr4_117131304_117132383 | 0.53 |
Plk3 |
polo like kinase 3 |
1956 |
0.12 |
| chr3_4796861_4798079 | 0.53 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
88 |
0.98 |
| chr7_90144986_90145137 | 0.53 |
Gm45222 |
predicted gene 45222 |
887 |
0.45 |
| chr10_123266080_123266231 | 0.53 |
Tafa2 |
TAFA chemokine like family member 2 |
1170 |
0.53 |
| chr12_72234504_72235243 | 0.53 |
Rtn1 |
reticulon 1 |
866 |
0.66 |
| chr18_43311052_43311203 | 0.52 |
Dpysl3 |
dihydropyrimidinase-like 3 |
34431 |
0.17 |
| chr3_58418728_58419191 | 0.52 |
Tsc22d2 |
TSC22 domain family, member 2 |
1475 |
0.4 |
| chr16_72693661_72693848 | 0.52 |
Robo1 |
roundabout guidance receptor 1 |
30550 |
0.26 |
| chr18_69354372_69354523 | 0.52 |
Tcf4 |
transcription factor 4 |
5503 |
0.29 |
| chr3_145591307_145591663 | 0.52 |
Znhit6 |
zinc finger, HIT type 6 |
4383 |
0.23 |
| chr9_80306711_80307237 | 0.52 |
Myo6 |
myosin VI |
60 |
0.98 |
| chr1_42709764_42710511 | 0.52 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
2085 |
0.24 |
| chr3_123688480_123688631 | 0.51 |
Gm42510 |
predicted gene 42510 |
583 |
0.69 |
| chr3_88534632_88537050 | 0.51 |
Mir1905 |
microRNA 1905 |
541 |
0.52 |
| chr16_38095027_38095536 | 0.50 |
Gsk3b |
glycogen synthase kinase 3 beta |
4993 |
0.25 |
| chr6_23836472_23836623 | 0.50 |
Cadps2 |
Ca2+-dependent activator protein for secretion 2 |
2590 |
0.4 |
| chr1_176811621_176811894 | 0.50 |
Cep170 |
centrosomal protein 170 |
2310 |
0.18 |
| chr17_40811481_40812037 | 0.49 |
Rhag |
Rhesus blood group-associated A glycoprotein |
575 |
0.7 |
| chr1_91543666_91543817 | 0.49 |
Asb1 |
ankyrin repeat and SOCS box-containing 1 |
2661 |
0.22 |
| chr4_132883220_132883371 | 0.49 |
Stx12 |
syntaxin 12 |
1214 |
0.3 |
| chr14_76432230_76432381 | 0.49 |
Tsc22d1 |
TSC22 domain family, member 1 |
5461 |
0.29 |
| chr11_98323740_98324755 | 0.49 |
Neurod2 |
neurogenic differentiation 2 |
5401 |
0.1 |
| chr9_41584760_41585051 | 0.48 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
2864 |
0.14 |
| chr17_47632404_47632673 | 0.48 |
Usp49 |
ubiquitin specific peptidase 49 |
1848 |
0.18 |
| chr3_88211471_88212561 | 0.48 |
Gm3764 |
predicted gene 3764 |
2469 |
0.11 |
| chrY_1285401_1285776 | 0.48 |
Ddx3y |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
994 |
0.48 |
| chr11_85341476_85341832 | 0.47 |
Bcas3 |
breast carcinoma amplified sequence 3 |
11513 |
0.2 |
| chr3_86448842_86449085 | 0.47 |
Gm25039 |
predicted gene, 25039 |
64716 |
0.1 |
| chr16_38293748_38294005 | 0.47 |
Nr1i2 |
nuclear receptor subfamily 1, group I, member 2 |
948 |
0.48 |
| chr1_61472065_61473256 | 0.47 |
Gm25839 |
predicted gene, 25839 |
2219 |
0.24 |
| chr4_15130212_15130363 | 0.47 |
Necab1 |
N-terminal EF-hand calcium binding protein 1 |
18674 |
0.21 |
| chr11_54026747_54027441 | 0.47 |
Slc22a4 |
solute carrier family 22 (organic cation transporter), member 4 |
858 |
0.54 |
| chr4_108849566_108849717 | 0.46 |
Kti12 |
KTI12 homolog, chromatin associated |
1856 |
0.25 |
| chr1_169929579_169930428 | 0.46 |
Ccdc190 |
coiled-coil domain containing 190 |
74 |
0.97 |
| chr3_17797523_17798047 | 0.46 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
2041 |
0.27 |
| chr17_71205988_71206421 | 0.46 |
Lpin2 |
lipin 2 |
1528 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 1.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.7 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 | 0.7 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 0.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.2 | 0.6 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.2 | 0.5 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.5 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.1 | 0.4 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.4 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.4 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.1 | GO:1901256 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.3 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.2 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 2.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.4 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.1 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.1 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 0.2 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.9 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 | 0.3 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.3 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 1.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0043738 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |