Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
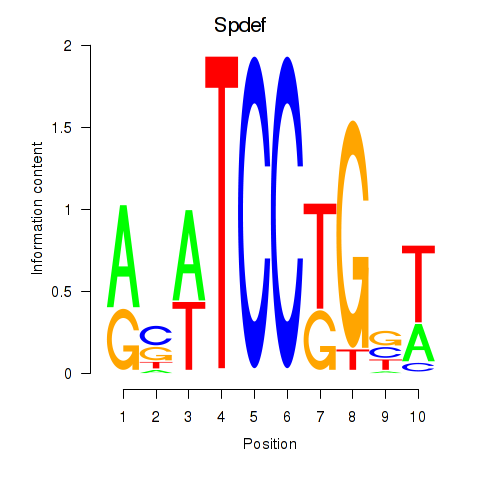
Results for Spdef
Z-value: 0.48
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.7 | SAM pointed domain containing ets transcription factor |
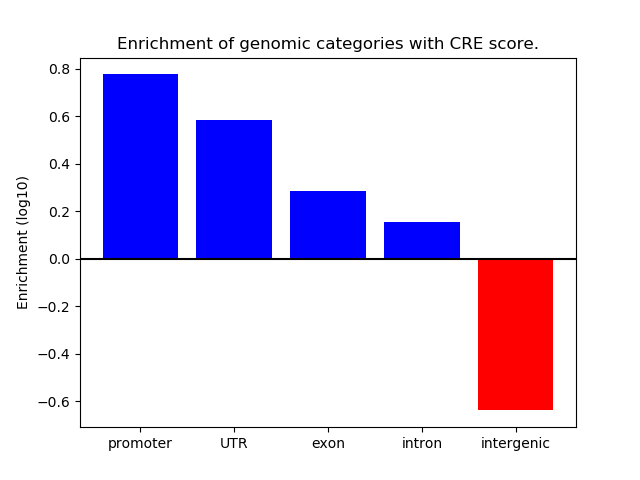
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_27726230_27726673 | Spdef | 2438 | 0.186996 | 0.41 | 1.3e-03 | Click! |
| chr17_27725174_27725943 | Spdef | 3331 | 0.155451 | 0.32 | 1.2e-02 | Click! |
| chr17_27726810_27726998 | Spdef | 1985 | 0.218732 | 0.28 | 3.3e-02 | Click! |
| chr17_27728247_27728914 | Spdef | 309 | 0.836350 | 0.19 | 1.4e-01 | Click! |
| chr17_27727948_27728145 | Spdef | 843 | 0.482827 | 0.12 | 3.5e-01 | Click! |
Activity of the Spdef motif across conditions
Conditions sorted by the z-value of the Spdef motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
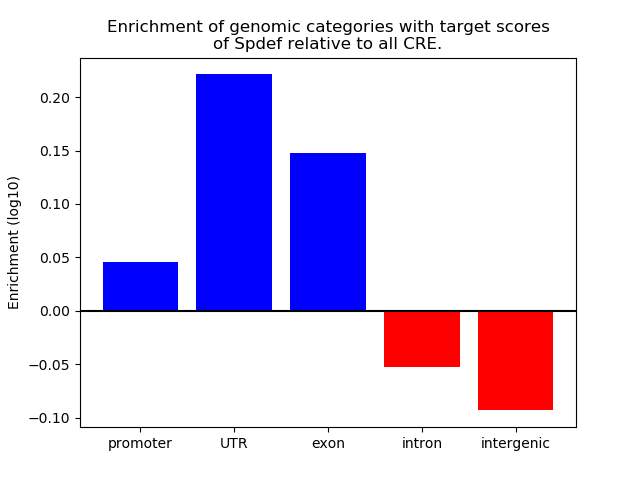
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_122459815_122460898 | 0.49 |
Zic5 |
zinc finger protein of the cerebellum 5 |
335 |
0.81 |
| chr19_12488341_12489388 | 0.46 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
12590 |
0.1 |
| chr7_16404362_16406481 | 0.46 |
Gm45508 |
predicted gene 45508 |
1159 |
0.29 |
| chr9_108824114_108825614 | 0.45 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr8_45509543_45510137 | 0.44 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
1922 |
0.33 |
| chr3_31097203_31098928 | 0.43 |
Skil |
SKI-like |
1232 |
0.48 |
| chr7_16982884_16983633 | 0.42 |
Gm42372 |
predicted gene, 42372 |
172 |
0.9 |
| chr14_122477737_122477888 | 0.41 |
Zic2 |
zinc finger protein of the cerebellum 2 |
288 |
0.83 |
| chrX_153501207_153502250 | 0.37 |
Ubqln2 |
ubiquilin 2 |
3501 |
0.22 |
| chrX_170676003_170677019 | 0.36 |
Asmt |
acetylserotonin O-methyltransferase |
3867 |
0.35 |
| chrX_105390628_105392456 | 0.34 |
5330434G04Rik |
RIKEN cDNA 5330434G04 gene |
212 |
0.93 |
| chr2_6883618_6884699 | 0.32 |
Gm13389 |
predicted gene 13389 |
112 |
0.85 |
| chr4_156183474_156184054 | 0.32 |
Agrn |
agrin |
2137 |
0.17 |
| chr10_92160735_92161461 | 0.31 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
1663 |
0.4 |
| chr5_120433178_120434996 | 0.30 |
Gm27199 |
predicted gene 27199 |
2320 |
0.19 |
| chr13_34127276_34127957 | 0.30 |
Tubb2b |
tubulin, beta 2B class IIB |
2738 |
0.15 |
| chr13_83715222_83716973 | 0.30 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
5284 |
0.15 |
| chr11_80479429_80480178 | 0.29 |
Cdk5r1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
2747 |
0.26 |
| chr1_6733530_6734383 | 0.29 |
St18 |
suppression of tumorigenicity 18 |
914 |
0.7 |
| chr11_96334278_96335532 | 0.28 |
Hoxb3 |
homeobox B3 |
6040 |
0.08 |
| chr4_110284419_110284817 | 0.28 |
Elavl4 |
ELAV like RNA binding protein 4 |
1998 |
0.47 |
| chr7_126041945_126042146 | 0.28 |
Gm44874 |
predicted gene 44874 |
22402 |
0.17 |
| chr8_89036575_89038609 | 0.27 |
Sall1 |
spalt like transcription factor 1 |
6570 |
0.23 |
| chr9_79829898_79830916 | 0.27 |
Filip1 |
filamin A interacting protein 1 |
591 |
0.7 |
| chr5_110543976_110545228 | 0.27 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
247 |
0.9 |
| chr4_6445169_6445688 | 0.27 |
Nsmaf |
neutral sphingomyelinase (N-SMase) activation associated factor |
899 |
0.62 |
| chr10_130279504_130280322 | 0.26 |
Neurod4 |
neurogenic differentiation 4 |
327 |
0.87 |
| chr5_73189782_73190392 | 0.26 |
Gm42571 |
predicted gene 42571 |
330 |
0.81 |
| chr9_44340460_44342952 | 0.26 |
Hmbs |
hydroxymethylbilane synthase |
473 |
0.51 |
| chr12_110187430_110189676 | 0.25 |
Gm34785 |
predicted gene, 34785 |
492 |
0.73 |
| chr10_79869256_79869624 | 0.25 |
Plppr3 |
phospholipid phosphatase related 3 |
2159 |
0.09 |
| chr1_77503797_77505059 | 0.25 |
Mir6352 |
microRNA 6352 |
7660 |
0.19 |
| chr13_83740463_83741042 | 0.25 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
1889 |
0.2 |
| chr2_151968839_151969749 | 0.24 |
Mir1953 |
microRNA 1953 |
1677 |
0.26 |
| chr5_139543263_139545139 | 0.24 |
Uncx |
UNC homeobox |
303 |
0.89 |
| chr13_99780787_99781532 | 0.24 |
Gm24471 |
predicted gene, 24471 |
88630 |
0.08 |
| chr11_69554884_69555035 | 0.23 |
Efnb3 |
ephrin B3 |
5246 |
0.09 |
| chr2_24473566_24475385 | 0.23 |
Pax8 |
paired box 8 |
622 |
0.64 |
| chr14_122475443_122476757 | 0.23 |
Zic2 |
zinc finger protein of the cerebellum 2 |
665 |
0.44 |
| chr2_74725879_74728683 | 0.23 |
Hoxd4 |
homeobox D4 |
207 |
0.67 |
| chr7_70108517_70108933 | 0.23 |
Gm35325 |
predicted gene, 35325 |
98710 |
0.07 |
| chr9_108829719_108830312 | 0.23 |
Celsr3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
1725 |
0.18 |
| chr7_133700065_133700317 | 0.23 |
Uros |
uroporphyrinogen III synthase |
2111 |
0.2 |
| chr4_124660613_124661391 | 0.23 |
Gm2164 |
predicted gene 2164 |
3833 |
0.13 |
| chr13_112695809_112696082 | 0.23 |
Gm18883 |
predicted gene, 18883 |
3734 |
0.17 |
| chr5_139546076_139547826 | 0.23 |
Uncx |
UNC homeobox |
3053 |
0.23 |
| chr11_96331807_96334248 | 0.22 |
Hoxb3 |
homeobox B3 |
4965 |
0.08 |
| chr2_18044333_18044941 | 0.22 |
Skida1 |
SKI/DACH domain containing 1 |
65 |
0.95 |
| chr2_157026864_157027931 | 0.22 |
Soga1 |
suppressor of glucose, autophagy associated 1 |
75 |
0.96 |
| chr19_53897189_53897340 | 0.22 |
Pdcd4 |
programmed cell death 4 |
3434 |
0.19 |
| chr7_126979881_126980032 | 0.22 |
Cdipt |
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
1723 |
0.13 |
| chr6_134886811_134888239 | 0.22 |
Gpr19 |
G protein-coupled receptor 19 |
243 |
0.87 |
| chr19_45193534_45193685 | 0.22 |
Tlx1os |
T cell leukemia, homeobox 1, opposite strand |
34305 |
0.12 |
| chr7_73378126_73378277 | 0.22 |
A730056A06Rik |
RIKEN cDNA A730056A06 gene |
2427 |
0.2 |
| chr1_177446374_177448525 | 0.22 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
1628 |
0.31 |
| chr14_77159311_77160431 | 0.22 |
Enox1 |
ecto-NOX disulfide-thiol exchanger 1 |
3091 |
0.31 |
| chr3_55782570_55784448 | 0.22 |
Nbea |
neurobeachin |
19 |
0.96 |
| chr1_132387629_132389330 | 0.22 |
Tmcc2 |
transmembrane and coiled-coil domains 2 |
1839 |
0.25 |
| chr2_151966993_151968089 | 0.22 |
Mir1953 |
microRNA 1953 |
76 |
0.96 |
| chr14_64588312_64589438 | 0.22 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
341 |
0.81 |
| chr10_109832096_109833441 | 0.22 |
Nav3 |
neuron navigator 3 |
453 |
0.9 |
| chr9_65555338_65555833 | 0.22 |
Plekho2 |
pleckstrin homology domain containing, family O member 2 |
6579 |
0.14 |
| chr6_90781027_90782541 | 0.21 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
188 |
0.94 |
| chr14_69269425_69270999 | 0.21 |
Gm27222 |
predicted gene 27222 |
11066 |
0.09 |
| chr6_28828230_28829085 | 0.21 |
Lrrc4 |
leucine rich repeat containing 4 |
1688 |
0.37 |
| chr2_146549745_146549896 | 0.21 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
6889 |
0.27 |
| chr1_115684558_115685809 | 0.21 |
Cntnap5a |
contactin associated protein-like 5A |
427 |
0.58 |
| chr4_154960570_154962200 | 0.21 |
Gm20421 |
predicted gene 20421 |
293 |
0.57 |
| chr8_122297355_122297506 | 0.21 |
Zfpm1 |
zinc finger protein, multitype 1 |
9890 |
0.15 |
| chr17_25930017_25930168 | 0.21 |
Pigq |
phosphatidylinositol glycan anchor biosynthesis, class Q |
916 |
0.29 |
| chr10_41070697_41071486 | 0.21 |
Gpr6 |
G protein-coupled receptor 6 |
1194 |
0.41 |
| chr1_66389272_66389745 | 0.21 |
Map2 |
microtubule-associated protein 2 |
2497 |
0.31 |
| chr13_37845693_37845844 | 0.21 |
Rreb1 |
ras responsive element binding protein 1 |
12166 |
0.19 |
| chr4_9269280_9270516 | 0.21 |
Clvs1 |
clavesin 1 |
551 |
0.81 |
| chr7_49915184_49916836 | 0.21 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
4310 |
0.29 |
| chr12_117157079_117158175 | 0.21 |
Gm10421 |
predicted gene 10421 |
5976 |
0.31 |
| chr15_59040592_59041370 | 0.21 |
Mtss1 |
MTSS I-BAR domain containing 1 |
384 |
0.88 |
| chr9_114404489_114404640 | 0.20 |
Glb1 |
galactosidase, beta 1 |
3456 |
0.17 |
| chr8_12399326_12400483 | 0.20 |
Gm25239 |
predicted gene, 25239 |
3501 |
0.16 |
| chr14_67320401_67320669 | 0.20 |
Gm6878 |
predicted gene 6878 |
5824 |
0.18 |
| chr9_91363064_91363717 | 0.20 |
Zic1 |
zinc finger protein of the cerebellum 1 |
336 |
0.72 |
| chr6_52160071_52161455 | 0.20 |
Hotairm1 |
Hoxa transcript antisense RNA, myeloid-specific 1 |
2239 |
0.09 |
| chr1_42691569_42692627 | 0.20 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
995 |
0.42 |
| chr3_152100022_152100538 | 0.20 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
2574 |
0.21 |
| chr2_165882235_165884653 | 0.20 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
766 |
0.54 |
| chr15_82334694_82334845 | 0.20 |
Naga |
N-acetyl galactosaminidase, alpha |
1196 |
0.22 |
| chr13_59669031_59669197 | 0.20 |
Golm1 |
golgi membrane protein 1 |
6658 |
0.1 |
| chr13_58812962_58813113 | 0.20 |
C630044B11Rik |
RIKEN cDNA C630044B11 gene |
884 |
0.48 |
| chr2_84386593_84388183 | 0.20 |
Calcrl |
calcitonin receptor-like |
12030 |
0.2 |
| chr11_101084072_101084686 | 0.20 |
Coasy |
Coenzyme A synthase |
484 |
0.59 |
| chr16_94092357_94092508 | 0.20 |
Sim2 |
single-minded family bHLH transcription factor 2 |
7501 |
0.15 |
| chr8_26844152_26844303 | 0.20 |
2310008N11Rik |
RIKEN cDNA 2310008N11 gene |
2228 |
0.31 |
| chr11_77947804_77948671 | 0.19 |
Sez6 |
seizure related gene 6 |
3068 |
0.17 |
| chr9_73115681_73115832 | 0.19 |
Rsl24d1 |
ribosomal L24 domain containing 1 |
1160 |
0.32 |
| chr11_84526864_84527823 | 0.19 |
Lhx1os |
LIM homeobox 1, opposite strand |
1659 |
0.31 |
| chr14_69493829_69494689 | 0.19 |
Gm37094 |
predicted gene, 37094 |
6131 |
0.11 |
| chr6_88803678_88803829 | 0.19 |
Gm44001 |
predicted gene, 44001 |
1510 |
0.26 |
| chr4_21685165_21686049 | 0.19 |
Prdm13 |
PR domain containing 13 |
174 |
0.95 |
| chr11_78496560_78498132 | 0.19 |
Sarm1 |
sterile alpha and HEAT/Armadillo motif containing 1 |
112 |
0.9 |
| chr13_21913780_21914888 | 0.19 |
Gm44456 |
predicted gene, 44456 |
10682 |
0.06 |
| chr14_69275580_69276440 | 0.19 |
Gm20236 |
predicted gene, 20236 |
6130 |
0.1 |
| chr15_75620417_75621249 | 0.19 |
Zfp41 |
zinc finger protein 41 |
878 |
0.44 |
| chr17_32808045_32808304 | 0.19 |
Zfp811 |
zinc finger protein 811 |
1668 |
0.15 |
| chr5_5507730_5508026 | 0.19 |
Cldn12 |
claudin 12 |
6571 |
0.17 |
| chr2_51144318_51145477 | 0.19 |
Rnd3 |
Rho family GTPase 3 |
4197 |
0.29 |
| chrX_13244980_13245286 | 0.19 |
2010308F09Rik |
RIKEN cDNA 2010308F09 gene |
2603 |
0.17 |
| chr11_77486623_77487566 | 0.19 |
Ankrd13b |
ankyrin repeat domain 13b |
2572 |
0.17 |
| chr14_118234662_118235031 | 0.19 |
Gm4675 |
predicted gene 4675 |
1386 |
0.29 |
| chr4_117148050_117148208 | 0.18 |
Gm13015 |
predicted gene 13015 |
794 |
0.3 |
| chr11_117281297_117281448 | 0.18 |
Septin9 |
septin 9 |
15126 |
0.17 |
| chr5_120431744_120433119 | 0.18 |
Gm27199 |
predicted gene 27199 |
664 |
0.41 |
| chr6_55453986_55454137 | 0.18 |
Adcyap1r1 |
adenylate cyclase activating polypeptide 1 receptor 1 |
1884 |
0.37 |
| chr15_75619003_75619950 | 0.18 |
Zfp41 |
zinc finger protein 41 |
2235 |
0.18 |
| chr16_23889696_23891150 | 0.18 |
Sst |
somatostatin |
535 |
0.77 |
| chr16_97292625_97293406 | 0.18 |
Bace2 |
beta-site APP-cleaving enzyme 2 |
63727 |
0.12 |
| chr2_68365331_68365864 | 0.18 |
Stk39 |
serine/threonine kinase 39 |
922 |
0.69 |
| chr15_25624832_25625841 | 0.18 |
Myo10 |
myosin X |
2787 |
0.25 |
| chr12_111690836_111691593 | 0.18 |
Gm36635 |
predicted gene, 36635 |
11868 |
0.09 |
| chr5_138278285_138278436 | 0.18 |
Gpc2 |
glypican 2 (cerebroglycan) |
123 |
0.87 |
| chr1_174175177_174176129 | 0.18 |
Spta1 |
spectrin alpha, erythrocytic 1 |
2877 |
0.14 |
| chr7_27735092_27735839 | 0.18 |
Zfp60 |
zinc finger protein 60 |
902 |
0.47 |
| chr7_103812275_103812465 | 0.18 |
Hbb-bt |
hemoglobin, beta adult t chain |
1626 |
0.15 |
| chr7_49911362_49912424 | 0.17 |
Slc6a5 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
193 |
0.96 |
| chr14_54567850_54568001 | 0.17 |
Ajuba |
ajuba LIM protein |
4157 |
0.1 |
| chr10_81114097_81114248 | 0.17 |
Map2k2 |
mitogen-activated protein kinase kinase 2 |
4440 |
0.08 |
| chr13_62885645_62885887 | 0.17 |
Fbp1 |
fructose bisphosphatase 1 |
2361 |
0.22 |
| chr10_19356598_19357962 | 0.17 |
Olig3 |
oligodendrocyte transcription factor 3 |
747 |
0.72 |
| chr7_119184137_119185474 | 0.17 |
Gpr139 |
G protein-coupled receptor 139 |
202 |
0.96 |
| chr7_121313033_121313561 | 0.17 |
4930486N12Rik |
RIKEN cDNA 4930486N12 gene |
53902 |
0.1 |
| chr5_98182267_98183697 | 0.17 |
Prdm8 |
PR domain containing 8 |
2004 |
0.26 |
| chr2_26207113_26207799 | 0.17 |
Gm27196 |
predicted gene 27196 |
754 |
0.37 |
| chr1_97540278_97540732 | 0.17 |
Gm37171 |
predicted gene, 37171 |
4943 |
0.29 |
| chr4_89691755_89691965 | 0.17 |
Dmrta1 |
doublesex and mab-3 related transcription factor like family A1 |
3662 |
0.36 |
| chr9_122595045_122595971 | 0.17 |
9530059O14Rik |
RIKEN cDNA 9530059O14 gene |
23006 |
0.13 |
| chr7_90886512_90887836 | 0.17 |
Gm45159 |
predicted gene 45159 |
102 |
0.88 |
| chr19_15339963_15340957 | 0.17 |
Gm24319 |
predicted gene, 24319 |
339544 |
0.01 |
| chrX_8072408_8072977 | 0.17 |
Suv39h1 |
suppressor of variegation 3-9 1 |
1566 |
0.17 |
| chr9_65144272_65144925 | 0.17 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
3386 |
0.18 |
| chr9_21557298_21557458 | 0.17 |
Carm1 |
coactivator-associated arginine methyltransferase 1 |
10220 |
0.1 |
| chr8_122613231_122613382 | 0.17 |
Trappc2l |
trafficking protein particle complex 2-like |
1660 |
0.17 |
| chr8_71680412_71681032 | 0.17 |
Jak3 |
Janus kinase 3 |
2387 |
0.13 |
| chr7_30331456_30331694 | 0.16 |
Gm50483 |
predicted gene, 50483 |
3223 |
0.09 |
| chr2_65563921_65564380 | 0.16 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
3342 |
0.29 |
| chr15_100402856_100403333 | 0.16 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
149 |
0.92 |
| chr16_18213980_18215144 | 0.16 |
4933432I09Rik |
RIKEN cDNA 4933432I09 gene |
699 |
0.36 |
| chr11_120825651_120826165 | 0.16 |
Fasn |
fatty acid synthase |
1361 |
0.25 |
| chr5_115945354_115946075 | 0.16 |
Cit |
citron |
417 |
0.82 |
| chr8_121300864_121302289 | 0.16 |
Gm26815 |
predicted gene, 26815 |
55260 |
0.11 |
| chr4_129261016_129261208 | 0.16 |
C77080 |
expressed sequence C77080 |
292 |
0.75 |
| chr8_104734057_104734499 | 0.16 |
Ces2a |
carboxylesterase 2A |
250 |
0.83 |
| chr8_105689654_105689805 | 0.16 |
Carmil2 |
capping protein regulator and myosin 1 linker 2 |
1177 |
0.25 |
| chr10_81212943_81213094 | 0.16 |
Atcay |
ataxia, cerebellar, Cayman type |
258 |
0.77 |
| chr5_115362856_115363022 | 0.16 |
4930401G09Rik |
RIKEN cDNA 4930401G09 gene |
2816 |
0.11 |
| chr15_80674911_80675205 | 0.16 |
Fam83f |
family with sequence similarity 83, member F |
3211 |
0.17 |
| chr12_29534253_29535510 | 0.16 |
Gm20208 |
predicted gene, 20208 |
10 |
0.8 |
| chr12_100998075_100998234 | 0.16 |
Gm36756 |
predicted gene, 36756 |
5182 |
0.14 |
| chr7_36704669_36704859 | 0.16 |
Gm37452 |
predicted gene, 37452 |
5410 |
0.14 |
| chr11_55600811_55601094 | 0.16 |
Glra1 |
glycine receptor, alpha 1 subunit |
6781 |
0.21 |
| chr13_54838015_54838197 | 0.16 |
Gm16250 |
predicted gene 16250 |
2798 |
0.2 |
| chr10_99267189_99269284 | 0.16 |
Gm48089 |
predicted gene, 48089 |
340 |
0.78 |
| chr8_123100430_123100798 | 0.16 |
Rpl13 |
ribosomal protein L13 |
1736 |
0.17 |
| chr4_119076605_119076792 | 0.16 |
Gm12866 |
predicted gene 12866 |
7587 |
0.12 |
| chr10_84457678_84458129 | 0.16 |
Gm47962 |
predicted gene, 47962 |
17077 |
0.12 |
| chr13_9114890_9115041 | 0.16 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
20983 |
0.14 |
| chr11_114793014_114793463 | 0.16 |
Btbd17 |
BTB (POZ) domain containing 17 |
2635 |
0.19 |
| chr14_28508967_28511864 | 0.16 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
203 |
0.89 |
| chrX_136822812_136823259 | 0.16 |
Plp1 |
proteolipid protein (myelin) 1 |
245 |
0.88 |
| chr1_193175099_193175314 | 0.16 |
A130010J15Rik |
RIKEN cDNA A130010J15 gene |
1462 |
0.25 |
| chr3_108410436_108412210 | 0.16 |
Celsr2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
4229 |
0.11 |
| chr11_54598816_54599372 | 0.16 |
Rapgef6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
2920 |
0.29 |
| chr19_53059154_53059713 | 0.16 |
1700054A03Rik |
RIKEN cDNA 1700054A03 gene |
16785 |
0.15 |
| chrX_53233797_53233998 | 0.16 |
Plac1 |
placental specific protein 1 |
6214 |
0.18 |
| chr7_80398560_80398838 | 0.15 |
Furin |
furin (paired basic amino acid cleaving enzyme) |
770 |
0.47 |
| chr16_20713164_20713474 | 0.15 |
Clcn2 |
chloride channel, voltage-sensitive 2 |
349 |
0.71 |
| chr2_65929929_65930575 | 0.15 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
115 |
0.97 |
| chr8_65966341_65967718 | 0.15 |
Marchf1 |
membrane associated ring-CH-type finger 1 |
128 |
0.97 |
| chr18_68398281_68398432 | 0.15 |
Mc2r |
melanocortin 2 receptor |
30964 |
0.15 |
| chr17_32334810_32334961 | 0.15 |
Akap8l |
A kinase (PRKA) anchor protein 8-like |
405 |
0.76 |
| chr12_11880981_11881577 | 0.15 |
Tubb2a-ps2 |
tubulin, beta 2a, pseudogene 2 |
1384 |
0.49 |
| chr15_98977285_98979178 | 0.15 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
5117 |
0.09 |
| chr9_55043837_55044704 | 0.15 |
Chrnb4 |
cholinergic receptor, nicotinic, beta polypeptide 4 |
4509 |
0.13 |
| chr4_155886040_155886274 | 0.15 |
Ints11 |
integrator complex subunit 11 |
1622 |
0.16 |
| chr5_115953777_115953928 | 0.15 |
Cit |
citron |
2762 |
0.23 |
| chr16_91685571_91685880 | 0.15 |
Donson |
downstream neighbor of SON |
1290 |
0.29 |
| chr2_167777077_167778323 | 0.15 |
Gm14321 |
predicted gene 14321 |
164 |
0.94 |
| chr7_99368824_99368975 | 0.15 |
Gdpd5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
12515 |
0.14 |
| chr10_128504713_128504957 | 0.15 |
Myl6b |
myosin, light polypeptide 6B |
6150 |
0.07 |
| chr3_17799177_17799545 | 0.15 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
3617 |
0.21 |
| chr5_136989551_136989702 | 0.15 |
Plod3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
680 |
0.41 |
| chr10_130276309_130276802 | 0.15 |
Neurod4 |
neurogenic differentiation 4 |
3685 |
0.2 |
| chr11_69399334_69402458 | 0.15 |
Kdm6bos |
KDM1 lysine (K)-specific demethylase 6B, opposite strand |
617 |
0.47 |
| chr8_110924044_110924195 | 0.15 |
Gm26132 |
predicted gene, 26132 |
1003 |
0.38 |
| chr8_23043503_23044627 | 0.15 |
Ank1 |
ankyrin 1, erythroid |
8834 |
0.18 |
| chr4_46255563_46255724 | 0.15 |
Gm12444 |
predicted gene 12444 |
11753 |
0.15 |
| chr1_132317853_132318004 | 0.15 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
1763 |
0.24 |
| chr11_118906363_118906514 | 0.15 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
1567 |
0.39 |
| chr13_99594358_99594644 | 0.15 |
Gm47158 |
predicted gene, 47158 |
33059 |
0.15 |
| chr1_75277211_75278430 | 0.15 |
Resp18 |
regulated endocrine-specific protein 18 |
464 |
0.62 |
| chr12_29537800_29538885 | 0.15 |
Myt1l |
myelin transcription factor 1-like |
3120 |
0.28 |
| chr10_6788358_6789268 | 0.15 |
Oprm1 |
opioid receptor, mu 1 |
4 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.2 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.1 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.2 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0097476 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.1 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.0 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.0 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.0 | GO:0042402 | cellular biogenic amine catabolic process(GO:0042402) |
| 0.0 | 0.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.0 | GO:0009310 | amine catabolic process(GO:0009310) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.0 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |