Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
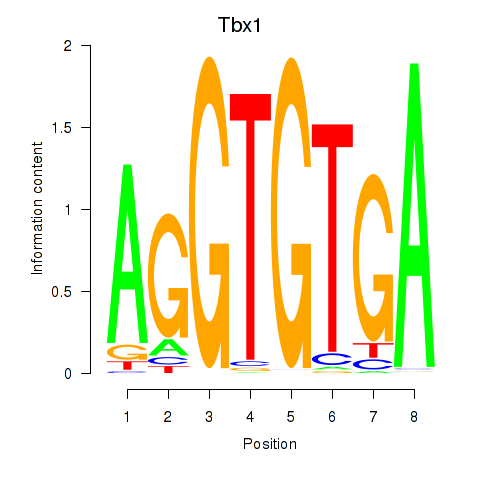
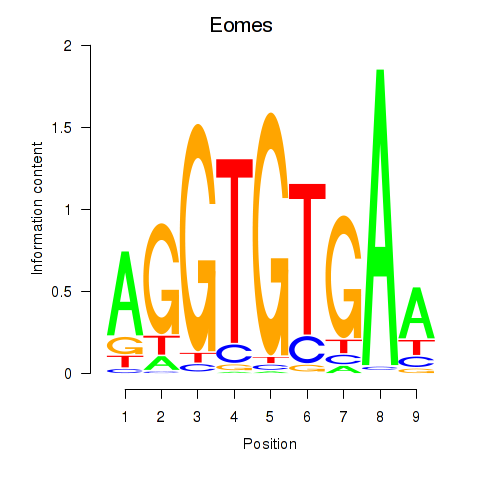
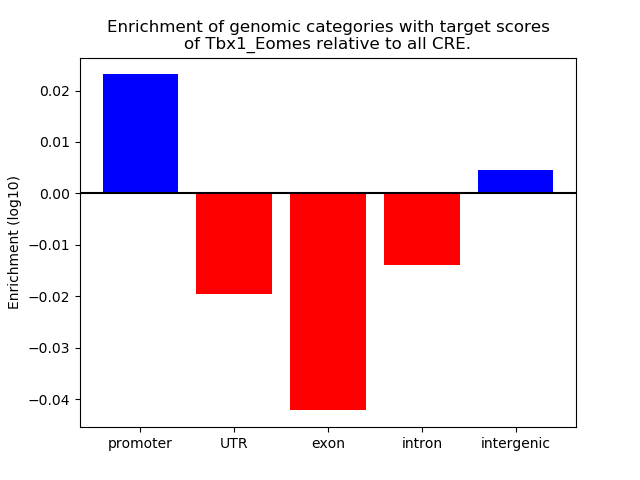
Results for Tbx1_Eomes
Z-value: 0.44
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx1
|
ENSMUSG00000009097.9 | T-box 1 |
|
Eomes
|
ENSMUSG00000032446.8 | eomesodermin |
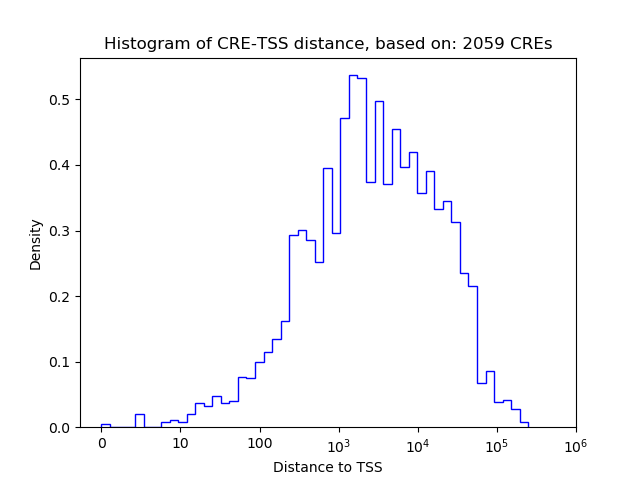
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_118476305_118476456 | Eomes | 1832 | 0.258459 | 0.27 | 4.0e-02 | Click! |
| chr9_118482050_118482392 | Eomes | 3360 | 0.172105 | -0.24 | 6.8e-02 | Click! |
| chr9_118476583_118478215 | Eomes | 813 | 0.531257 | 0.24 | 6.9e-02 | Click! |
| chr9_118478372_118479858 | Eomes | 254 | 0.887315 | 0.19 | 1.4e-01 | Click! |
| chr9_118481236_118481395 | Eomes | 2454 | 0.206561 | -0.17 | 1.8e-01 | Click! |
| chr16_18589901_18590901 | Tbx1 | 270 | 0.835432 | 0.53 | 1.7e-05 | Click! |
| chr16_18587715_18589011 | Tbx1 | 445 | 0.727351 | 0.51 | 3.7e-05 | Click! |
| chr16_18585593_18587306 | Tbx1 | 520 | 0.677503 | 0.46 | 2.1e-04 | Click! |
| chr16_18585265_18585440 | Tbx1 | 1617 | 0.259728 | 0.40 | 1.7e-03 | Click! |
| chr16_18589340_18589872 | Tbx1 | 798 | 0.479543 | 0.33 | 9.6e-03 | Click! |
Activity of the Tbx1_Eomes motif across conditions
Conditions sorted by the z-value of the Tbx1_Eomes motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
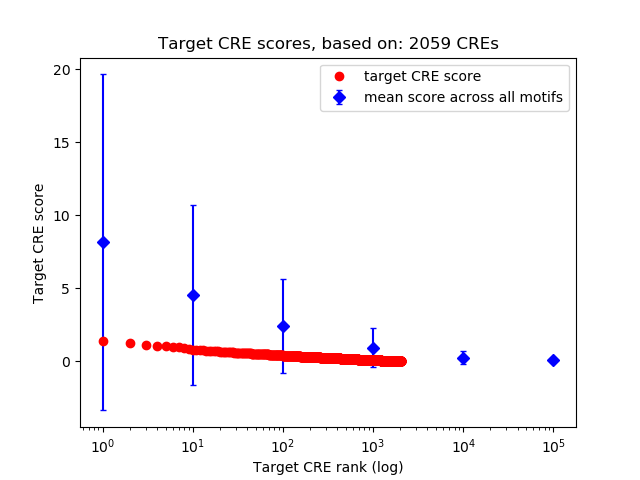
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_99709458_99709950 | 1.42 |
Cldn18 |
claudin 18 |
305 |
0.84 |
| chr3_19646421_19646748 | 1.27 |
Trim55 |
tripartite motif-containing 55 |
2076 |
0.26 |
| chr2_163396102_163396417 | 1.11 |
Jph2 |
junctophilin 2 |
1690 |
0.28 |
| chr2_91120847_91121416 | 1.05 |
Mybpc3 |
myosin binding protein C, cardiac |
2987 |
0.16 |
| chr2_163395773_163396078 | 1.03 |
Jph2 |
junctophilin 2 |
2024 |
0.24 |
| chr1_34116331_34116653 | 0.99 |
Mir6896 |
microRNA 6896 |
867 |
0.57 |
| chr8_61901105_61901625 | 0.97 |
Palld |
palladin, cytoskeletal associated protein |
1304 |
0.42 |
| chr5_134745620_134746234 | 0.88 |
Gm30003 |
predicted gene, 30003 |
1115 |
0.35 |
| chr3_155054087_155054557 | 0.82 |
Tnni3k |
TNNI3 interacting kinase |
1028 |
0.55 |
| chr2_32318746_32319267 | 0.79 |
Mir199b |
microRNA 199b |
546 |
0.42 |
| chr13_23913630_23913830 | 0.78 |
Slc17a4 |
solute carrier family 17 (sodium phosphate), member 4 |
1238 |
0.27 |
| chr17_81737002_81738450 | 0.75 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
651 |
0.81 |
| chr2_92140355_92141595 | 0.75 |
Phf21a |
PHD finger protein 21A |
43131 |
0.11 |
| chr15_12010211_12011091 | 0.74 |
Sub1 |
SUB1 homolog, transcriptional regulator |
13668 |
0.16 |
| chr13_63241575_63242090 | 0.72 |
Aopep |
aminopeptidase O |
1724 |
0.2 |
| chr15_3580312_3581004 | 0.70 |
Ghr |
growth hormone receptor |
1184 |
0.57 |
| chr16_87553258_87554074 | 0.69 |
Map3k7cl |
Map3k7 C-terminal like |
336 |
0.84 |
| chr1_51291546_51292409 | 0.68 |
Cavin2 |
caveolae associated 2 |
2851 |
0.27 |
| chr4_141302549_141302700 | 0.68 |
Epha2 |
Eph receptor A2 |
217 |
0.88 |
| chrX_152768117_152768457 | 0.66 |
Shroom2 |
shroom family member 2 |
1174 |
0.51 |
| chr2_166412306_166412566 | 0.64 |
Gm14268 |
predicted gene 14268 |
14249 |
0.21 |
| chr9_44069968_44070511 | 0.64 |
Usp2 |
ubiquitin specific peptidase 2 |
796 |
0.33 |
| chr5_75152837_75154692 | 0.64 |
Gm42802 |
predicted gene 42802 |
111 |
0.58 |
| chr16_36989294_36990265 | 0.64 |
Fbxo40 |
F-box protein 40 |
688 |
0.6 |
| chr8_27261786_27261937 | 0.63 |
Eif4ebp1 |
eukaryotic translation initiation factor 4E binding protein 1 |
1532 |
0.26 |
| chr8_25027794_25028434 | 0.63 |
Gm16933 |
predicted gene, 16933 |
8938 |
0.12 |
| chr18_35120289_35121263 | 0.61 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
1334 |
0.45 |
| chr9_45934543_45935100 | 0.61 |
Tagln |
transgelin |
753 |
0.43 |
| chr16_44015370_44016774 | 0.59 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr10_22470665_22470816 | 0.58 |
Gm26585 |
predicted gene, 26585 |
17220 |
0.18 |
| chr10_120900758_120900909 | 0.58 |
Gm4473 |
predicted gene 4473 |
1676 |
0.25 |
| chr3_30546274_30547260 | 0.58 |
Mecom |
MDS1 and EVI1 complex locus |
1241 |
0.35 |
| chr3_49753928_49755610 | 0.58 |
Pcdh18 |
protocadherin 18 |
695 |
0.72 |
| chr7_130576739_130577783 | 0.58 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
177 |
0.94 |
| chr12_82122852_82123243 | 0.57 |
n-R5s62 |
nuclear encoded rRNA 5S 62 |
27421 |
0.18 |
| chr18_53466213_53466522 | 0.57 |
Prdm6 |
PR domain containing 6 |
1851 |
0.45 |
| chr5_100844231_100844692 | 0.57 |
Gpat3 |
glycerol-3-phosphate acyltransferase 3 |
1252 |
0.27 |
| chr9_119962629_119963694 | 0.56 |
Ttc21a |
tetratricopeptide repeat domain 21A |
1543 |
0.24 |
| chr10_22471094_22471245 | 0.56 |
Gm26585 |
predicted gene, 26585 |
17649 |
0.18 |
| chr19_56120628_56120882 | 0.55 |
Gm31912 |
predicted gene, 31912 |
14445 |
0.24 |
| chr8_41040672_41042201 | 0.55 |
Mtus1 |
mitochondrial tumor suppressor 1 |
430 |
0.76 |
| chr6_71254405_71254608 | 0.55 |
Smyd1 |
SET and MYND domain containing 1 |
7726 |
0.11 |
| chr14_63265100_63265475 | 0.54 |
Gata4 |
GATA binding protein 4 |
5837 |
0.19 |
| chr11_97670192_97670699 | 0.54 |
Mllt6 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
5444 |
0.08 |
| chr3_53043050_53043701 | 0.53 |
Gm42901 |
predicted gene 42901 |
1620 |
0.23 |
| chr15_79111462_79112012 | 0.53 |
Micall1 |
microtubule associated monooxygenase, calponin and LIM domain containing -like 1 |
2407 |
0.14 |
| chr13_69460904_69461114 | 0.53 |
Gm48676 |
predicted gene, 48676 |
52991 |
0.09 |
| chr10_116750858_116751292 | 0.52 |
4930579P08Rik |
RIKEN cDNA 4930579P08 gene |
21822 |
0.16 |
| chr3_89832920_89834058 | 0.52 |
She |
src homology 2 domain-containing transforming protein E |
2119 |
0.21 |
| chr5_119742473_119742643 | 0.52 |
Tbx3os2 |
T-box 3, opposite strand 2 |
51340 |
0.1 |
| chr1_74748322_74749490 | 0.52 |
Prkag3 |
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
2 |
0.96 |
| chr7_16892200_16892882 | 0.52 |
Gng8 |
guanine nucleotide binding protein (G protein), gamma 8 |
220 |
0.83 |
| chr6_129183729_129184848 | 0.51 |
Clec2d |
C-type lectin domain family 2, member d |
3673 |
0.14 |
| chr5_137349923_137351747 | 0.51 |
Ephb4 |
Eph receptor B4 |
322 |
0.77 |
| chr11_103096614_103097909 | 0.51 |
Plcd3 |
phospholipase C, delta 3 |
4330 |
0.13 |
| chr3_132948328_132948925 | 0.51 |
Npnt |
nephronectin |
1188 |
0.43 |
| chr13_46423537_46423785 | 0.51 |
Rbm24 |
RNA binding motif protein 24 |
1839 |
0.41 |
| chr19_53557067_53557881 | 0.51 |
Gm50394 |
predicted gene, 50394 |
27819 |
0.11 |
| chr2_35623965_35624326 | 0.50 |
Dab2ip |
disabled 2 interacting protein |
1985 |
0.37 |
| chr14_58428834_58429409 | 0.50 |
Gm33321 |
predicted gene, 33321 |
128139 |
0.05 |
| chr8_57324709_57326732 | 0.49 |
Hand2os1 |
Hand2, opposite strand 1 |
1487 |
0.3 |
| chr9_79990471_79990814 | 0.49 |
Filip1 |
filamin A interacting protein 1 |
12838 |
0.16 |
| chr2_35588919_35589070 | 0.48 |
Dab2ip |
disabled 2 interacting protein |
6154 |
0.2 |
| chr14_74852987_74853831 | 0.48 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
31846 |
0.18 |
| chr6_4005591_4005747 | 0.48 |
Gng11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1765 |
0.31 |
| chr6_124925245_124926316 | 0.48 |
Mlf2 |
myeloid leukemia factor 2 |
5606 |
0.08 |
| chr4_62755430_62755581 | 0.47 |
Gm11211 |
predicted gene 11211 |
27968 |
0.15 |
| chr18_75279035_75279335 | 0.47 |
2010010A06Rik |
RIKEN cDNA 2010010A06 gene |
9998 |
0.23 |
| chr11_74526613_74527288 | 0.47 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
4199 |
0.25 |
| chr14_25323823_25323974 | 0.46 |
Gm26660 |
predicted gene, 26660 |
54737 |
0.12 |
| chr15_31396603_31396758 | 0.46 |
Gm49300 |
predicted gene, 49300 |
28318 |
0.11 |
| chr10_80960844_80961038 | 0.46 |
Gm3828 |
predicted gene 3828 |
6231 |
0.1 |
| chr12_84643018_84643308 | 0.46 |
Vrtn |
vertebrae development associated |
267 |
0.89 |
| chr4_133040822_133040973 | 0.46 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
1404 |
0.43 |
| chr14_26439572_26439816 | 0.45 |
Slmap |
sarcolemma associated protein |
473 |
0.78 |
| chr8_57340613_57341952 | 0.45 |
5033428I22Rik |
RIKEN cDNA 5033428I22 gene |
482 |
0.73 |
| chr16_77569814_77569965 | 0.45 |
Gm37606 |
predicted gene, 37606 |
8239 |
0.11 |
| chr6_48839906_48841160 | 0.44 |
Tmem176b |
transmembrane protein 176B |
62 |
0.82 |
| chr3_38015645_38016389 | 0.44 |
Gm22899 |
predicted gene, 22899 |
20782 |
0.16 |
| chr17_86754057_86754608 | 0.44 |
Epas1 |
endothelial PAS domain protein 1 |
632 |
0.71 |
| chr5_140890731_140891173 | 0.44 |
Card11 |
caspase recruitment domain family, member 11 |
8747 |
0.24 |
| chr13_46424277_46424435 | 0.44 |
Rbm24 |
RNA binding motif protein 24 |
2534 |
0.34 |
| chr15_83526325_83527694 | 0.43 |
Bik |
BCL2-interacting killer |
137 |
0.93 |
| chr18_3274616_3274842 | 0.43 |
Crem |
cAMP responsive element modulator |
6349 |
0.23 |
| chr15_89137295_89137649 | 0.43 |
Mapk12 |
mitogen-activated protein kinase 12 |
28 |
0.94 |
| chr13_102036243_102036435 | 0.43 |
Gm38133 |
predicted gene, 38133 |
79696 |
0.1 |
| chr6_52965535_52965952 | 0.43 |
Gm26215 |
predicted gene, 26215 |
44789 |
0.14 |
| chr19_56507368_56507519 | 0.42 |
Plekhs1 |
pleckstrin homology domain containing, family S member 1 |
27756 |
0.15 |
| chr7_19163884_19165719 | 0.42 |
Gipr |
gastric inhibitory polypeptide receptor |
293 |
0.77 |
| chr12_103336858_103338251 | 0.42 |
Gm15523 |
predicted gene 15523 |
648 |
0.43 |
| chr5_144507108_144507703 | 0.42 |
Nptx2 |
neuronal pentraxin 2 |
38497 |
0.17 |
| chr19_46448230_46448462 | 0.42 |
Sufu |
SUFU negative regulator of hedgehog signaling |
9560 |
0.15 |
| chr3_60504423_60505028 | 0.42 |
Mbnl1 |
muscleblind like splicing factor 1 |
3447 |
0.29 |
| chr14_118385532_118385782 | 0.42 |
Gm5672 |
predicted gene 5672 |
9463 |
0.16 |
| chr16_58685640_58686063 | 0.41 |
Cpox |
coproporphyrinogen oxidase |
7995 |
0.13 |
| chr5_75046578_75047197 | 0.40 |
Gm18593 |
predicted gene, 18593 |
1592 |
0.25 |
| chr5_32990256_32991187 | 0.40 |
Gm43693 |
predicted gene 43693 |
6151 |
0.16 |
| chr5_53274000_53274222 | 0.40 |
Smim20 |
small integral membrane protein 20 |
3007 |
0.24 |
| chr14_36917800_36917970 | 0.40 |
Ccser2 |
coiled-coil serine rich 2 |
1448 |
0.48 |
| chr18_84089861_84090362 | 0.39 |
Zadh2 |
zinc binding alcohol dehydrogenase, domain containing 2 |
1440 |
0.33 |
| chr6_112728907_112729181 | 0.39 |
Rad18 |
RAD18 E3 ubiquitin protein ligase |
32358 |
0.16 |
| chr3_32822040_32822406 | 0.39 |
Usp13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
4512 |
0.22 |
| chr11_18870055_18872175 | 0.39 |
8430419K02Rik |
RIKEN cDNA 8430419K02 gene |
1040 |
0.49 |
| chr2_26585065_26586013 | 0.39 |
Egfl7 |
EGF-like domain 7 |
1091 |
0.24 |
| chr17_69120419_69121537 | 0.39 |
Epb41l3 |
erythrocyte membrane protein band 4.1 like 3 |
6022 |
0.31 |
| chr4_154630838_154632339 | 0.39 |
Prdm16 |
PR domain containing 16 |
5209 |
0.14 |
| chr4_43763346_43763579 | 0.39 |
Olfr269-ps1 |
olfactory receptor 269, pseudogene 1 |
4315 |
0.1 |
| chr7_100657972_100658903 | 0.38 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
15 |
0.96 |
| chr11_54962647_54963034 | 0.38 |
Tnip1 |
TNFAIP3 interacting protein 1 |
69 |
0.97 |
| chr8_126587716_126588216 | 0.38 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
6020 |
0.24 |
| chr18_54452409_54453689 | 0.38 |
Gm50361 |
predicted gene, 50361 |
23716 |
0.19 |
| chr10_118839718_118840226 | 0.38 |
Gm47434 |
predicted gene, 47434 |
4866 |
0.15 |
| chr9_120619342_120620352 | 0.38 |
Gm39456 |
predicted gene, 39456 |
452 |
0.76 |
| chr6_24721472_24721728 | 0.37 |
Hyal6 |
hyaluronoglucosaminidase 6 |
11645 |
0.17 |
| chr12_31270509_31272027 | 0.37 |
Lamb1 |
laminin B1 |
5388 |
0.14 |
| chr4_83048506_83049199 | 0.37 |
Frem1 |
Fras1 related extracellular matrix protein 1 |
3315 |
0.28 |
| chr9_109065033_109065184 | 0.37 |
Atrip |
ATR interacting protein |
285 |
0.8 |
| chr15_103383151_103383913 | 0.37 |
Gm37322 |
predicted gene, 37322 |
5461 |
0.13 |
| chr10_61906333_61906511 | 0.37 |
Col13a1 |
collagen, type XIII, alpha 1 |
28030 |
0.17 |
| chr1_153329741_153330128 | 0.37 |
Lamc1 |
laminin, gamma 1 |
2852 |
0.24 |
| chr5_73231934_73232445 | 0.37 |
Gm17207 |
predicted gene 17207 |
19546 |
0.1 |
| chr11_95841518_95842457 | 0.37 |
Abi3 |
ABI gene family, member 3 |
111 |
0.73 |
| chr2_156717216_156718150 | 0.37 |
Dlgap4 |
DLG associated protein 4 |
2889 |
0.18 |
| chr4_9606607_9606758 | 0.37 |
Asph |
aspartate-beta-hydroxylase |
10993 |
0.25 |
| chr17_93437013_93437164 | 0.37 |
Gm50001 |
predicted gene, 50001 |
45390 |
0.17 |
| chr8_124943393_124943545 | 0.37 |
Egln1 |
egl-9 family hypoxia-inducible factor 1 |
5855 |
0.15 |
| chr12_83770139_83771096 | 0.36 |
Papln |
papilin, proteoglycan-like sulfated glycoprotein |
6955 |
0.12 |
| chr8_84558613_84558764 | 0.36 |
Cacna1a |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
11594 |
0.17 |
| chr1_189782721_189783385 | 0.36 |
Ptpn14 |
protein tyrosine phosphatase, non-receptor type 14 |
3583 |
0.24 |
| chr8_69023381_69023534 | 0.36 |
Gm15716 |
predicted gene 15716 |
21297 |
0.15 |
| chr1_165706603_165708435 | 0.36 |
Rcsd1 |
RCSD domain containing 1 |
529 |
0.64 |
| chr2_32311532_32312331 | 0.36 |
Dnm1 |
dynamin 1 |
958 |
0.33 |
| chr8_33916300_33917561 | 0.36 |
Rbpms |
RNA binding protein gene with multiple splicing |
12346 |
0.17 |
| chr14_49244860_49245366 | 0.36 |
ccdc198 |
coiled-coil domain containing 198 |
284 |
0.89 |
| chr6_22297902_22298203 | 0.36 |
Wnt16 |
wingless-type MMTV integration site family, member 16 |
9191 |
0.23 |
| chr15_101411658_101413570 | 0.35 |
Krt7 |
keratin 7 |
165 |
0.64 |
| chr17_78676930_78677295 | 0.35 |
Strn |
striatin, calmodulin binding protein |
7240 |
0.21 |
| chr5_149312765_149313642 | 0.35 |
Gm19719 |
predicted gene, 19719 |
25335 |
0.09 |
| chr15_51854075_51854251 | 0.35 |
Eif3h |
eukaryotic translation initiation factor 3, subunit H |
11291 |
0.17 |
| chr6_48647599_48648258 | 0.35 |
Gimap8 |
GTPase, IMAP family member 8 |
544 |
0.53 |
| chr11_103101106_103101632 | 0.35 |
Plcd3 |
phospholipase C, delta 3 |
222 |
0.58 |
| chr2_181241290_181242534 | 0.35 |
Helz2 |
helicase with zinc finger 2, transcriptional coactivator |
51 |
0.95 |
| chr2_91125839_91126091 | 0.35 |
Mybpc3 |
myosin binding protein C, cardiac |
7821 |
0.12 |
| chr4_137476144_137477534 | 0.35 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
8036 |
0.14 |
| chr3_98245804_98245958 | 0.34 |
Gm42821 |
predicted gene 42821 |
2718 |
0.21 |
| chr13_41345830_41346374 | 0.34 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
13145 |
0.14 |
| chr1_98132044_98132246 | 0.34 |
1810006J02Rik |
RIKEN cDNA 1810006J02 gene |
678 |
0.7 |
| chr5_147722674_147723044 | 0.34 |
Flt1 |
FMS-like tyrosine kinase 1 |
3129 |
0.27 |
| chr12_110504246_110504860 | 0.34 |
Gm19605 |
predicted gene, 19605 |
18345 |
0.15 |
| chr5_100214874_100215472 | 0.34 |
2310034O05Rik |
RIKEN cDNA 2310034O05 gene |
4465 |
0.21 |
| chr5_147723576_147724353 | 0.34 |
Flt1 |
FMS-like tyrosine kinase 1 |
2024 |
0.35 |
| chr10_22813360_22814229 | 0.34 |
Gm10824 |
predicted gene 10824 |
1955 |
0.28 |
| chr18_68058494_68058645 | 0.34 |
Gm41764 |
predicted gene, 41764 |
59805 |
0.12 |
| chr3_82141940_82142375 | 0.33 |
Gucy1a1 |
guanylate cyclase 1, soluble, alpha 1 |
2916 |
0.3 |
| chr1_38629122_38629273 | 0.33 |
Aff3 |
AF4/FMR2 family, member 3 |
1996 |
0.42 |
| chr8_54571415_54571982 | 0.33 |
Asb5 |
ankyrin repeat and SOCs box-containing 5 |
272 |
0.9 |
| chr17_34580979_34581380 | 0.33 |
Notch4 |
notch 4 |
2703 |
0.08 |
| chr17_35725752_35726163 | 0.33 |
Gm20443 |
predicted gene 20443 |
13849 |
0.08 |
| chr4_19576297_19576523 | 0.32 |
Rmdn1 |
regulator of microtubule dynamics 1 |
1203 |
0.48 |
| chr6_15638549_15638700 | 0.32 |
Gm44039 |
predicted gene, 44039 |
2500 |
0.4 |
| chr6_115984719_115988278 | 0.32 |
Plxnd1 |
plexin D1 |
8507 |
0.15 |
| chr13_93307758_93308718 | 0.32 |
Homer1 |
homer scaffolding protein 1 |
204 |
0.92 |
| chr12_56298063_56298592 | 0.32 |
Gm47682 |
predicted gene, 47682 |
43929 |
0.1 |
| chr4_45896655_45896836 | 0.32 |
Ccdc180 |
coiled-coil domain containing 180 |
5893 |
0.19 |
| chr14_101510884_101511350 | 0.32 |
Tbc1d4 |
TBC1 domain family, member 4 |
3791 |
0.3 |
| chr18_53249928_53250416 | 0.32 |
Snx24 |
sorting nexing 24 |
4403 |
0.25 |
| chr12_84873778_84874595 | 0.32 |
D030025P21Rik |
RIKEN cDNA D030025P21 gene |
1583 |
0.26 |
| chr9_111119102_111119253 | 0.32 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
252 |
0.9 |
| chr9_123480978_123481680 | 0.32 |
Limd1 |
LIM domains containing 1 |
719 |
0.66 |
| chr7_4515813_4516413 | 0.32 |
Tnnt1 |
troponin T1, skeletal, slow |
109 |
0.91 |
| chr18_61785451_61786937 | 0.32 |
Afap1l1 |
actin filament associated protein 1-like 1 |
417 |
0.83 |
| chr15_102249163_102250297 | 0.32 |
Rarg |
retinoic acid receptor, gamma |
616 |
0.54 |
| chr5_101239366_101239752 | 0.31 |
Cycs-ps2 |
cytochrome c, pseudogene 2 |
66019 |
0.13 |
| chr1_97975537_97975875 | 0.31 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
1362 |
0.45 |
| chr16_85546798_85547372 | 0.31 |
Cyyr1 |
cysteine and tyrosine-rich protein 1 |
3332 |
0.28 |
| chr11_108758258_108758409 | 0.31 |
Cep112 |
centrosomal protein 112 |
6128 |
0.22 |
| chr4_9184182_9184333 | 0.31 |
Gm23423 |
predicted gene, 23423 |
25746 |
0.23 |
| chr11_99042414_99042858 | 0.31 |
Igfbp4 |
insulin-like growth factor binding protein 4 |
450 |
0.73 |
| chr10_18432775_18433290 | 0.31 |
Nhsl1 |
NHS-like 1 |
25357 |
0.21 |
| chr3_95991778_95991929 | 0.31 |
Plekho1 |
pleckstrin homology domain containing, family O member 1 |
3815 |
0.14 |
| chr7_19011655_19012657 | 0.31 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
8112 |
0.07 |
| chr2_148037835_148038211 | 0.31 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
247 |
0.91 |
| chr6_99023814_99025127 | 0.31 |
Foxp1 |
forkhead box P1 |
3521 |
0.33 |
| chr10_13092588_13092795 | 0.31 |
Plagl1 |
pleiomorphic adenoma gene-like 1 |
1678 |
0.4 |
| chr14_69053224_69053475 | 0.31 |
Gm41192 |
predicted gene, 41192 |
23697 |
0.16 |
| chr9_53935727_53936526 | 0.31 |
Elmod1 |
ELMO/CED-12 domain containing 1 |
708 |
0.71 |
| chr11_114478869_114479221 | 0.31 |
4932435O22Rik |
RIKEN cDNA 4932435O22 gene |
30185 |
0.2 |
| chr8_121130074_121130554 | 0.30 |
Foxl1 |
forkhead box L1 |
2374 |
0.2 |
| chr2_144676648_144676927 | 0.30 |
Dtd1 |
D-tyrosyl-tRNA deacylase 1 |
1311 |
0.47 |
| chr1_188011031_188011182 | 0.30 |
9330162B11Rik |
RIKEN cDNA 9330162B11 gene |
2116 |
0.38 |
| chr2_145787122_145787273 | 0.30 |
Rin2 |
Ras and Rab interactor 2 |
1035 |
0.61 |
| chr19_20725054_20725278 | 0.30 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
2396 |
0.39 |
| chr15_103182225_103182894 | 0.30 |
Smug1 |
single-strand selective monofunctional uracil DNA glycosylase |
15467 |
0.11 |
| chr17_66404980_66405904 | 0.30 |
Gm4705 |
predicted gene 4705 |
8542 |
0.17 |
| chr6_51173127_51174365 | 0.30 |
Mir148a |
microRNA 148a |
96164 |
0.07 |
| chr2_35627849_35628114 | 0.29 |
Dab2ip |
disabled 2 interacting protein |
5821 |
0.24 |
| chr1_19241815_19242084 | 0.29 |
Gm28340 |
predicted gene 28340 |
776 |
0.67 |
| chr11_53132369_53132523 | 0.29 |
Gm11186 |
predicted gene 11186 |
12390 |
0.23 |
| chr12_28998072_28998252 | 0.29 |
Gm23342 |
predicted gene, 23342 |
37132 |
0.17 |
| chr19_44726477_44726822 | 0.29 |
Gm35610 |
predicted gene, 35610 |
13230 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.6 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.5 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.4 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.3 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.4 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.1 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.2 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.2 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.0 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.4 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.0 | 0.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0098911 | regulation of ventricular cardiac muscle cell action potential(GO:0098911) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.0 | GO:1903140 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.0 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.0 | GO:2001198 | regulation of dendritic cell differentiation(GO:2001198) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.0 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.0 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.0 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.0 | GO:1902990 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.4 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0046977 | TAP binding(GO:0046977) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |