Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
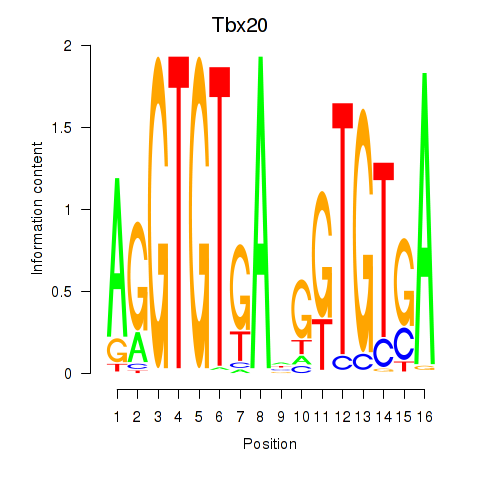
Results for Tbx20
Z-value: 0.68
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSMUSG00000031965.8 | T-box 20 |
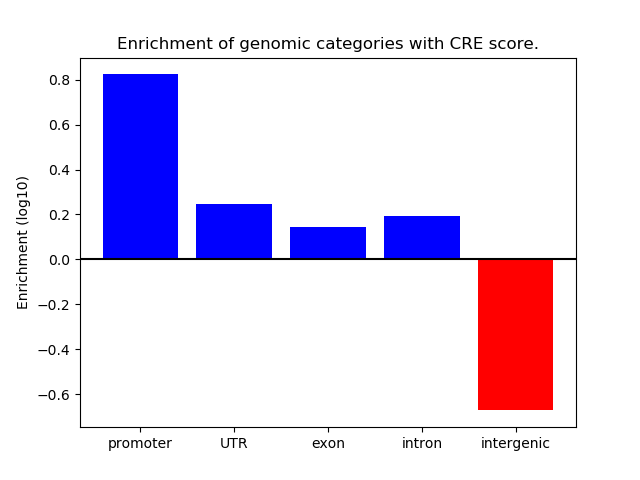
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_24750162_24750521 | Tbx20 | 19339 | 0.172676 | 0.37 | 3.2e-03 | Click! |
| chr9_24739789_24739940 | Tbx20 | 29816 | 0.151744 | 0.34 | 7.9e-03 | Click! |
| chr9_24768577_24769049 | Tbx20 | 867 | 0.602051 | 0.33 | 9.2e-03 | Click! |
| chr9_24769617_24771807 | Tbx20 | 962 | 0.556580 | 0.32 | 1.1e-02 | Click! |
| chr9_24773249_24774873 | Tbx20 | 223 | 0.923444 | 0.30 | 1.8e-02 | Click! |
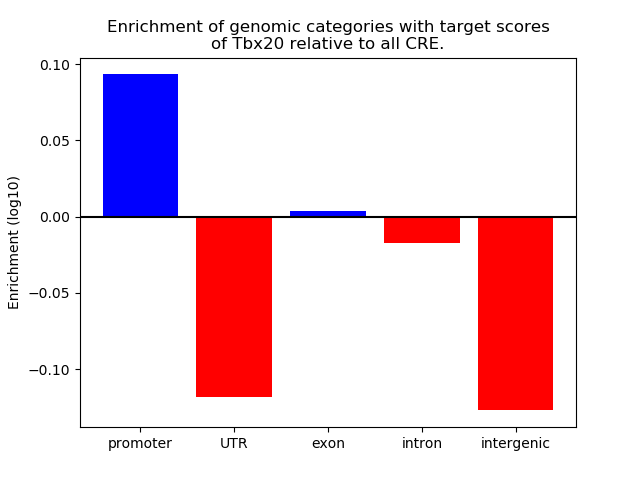
Activity of the Tbx20 motif across conditions
Conditions sorted by the z-value of the Tbx20 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
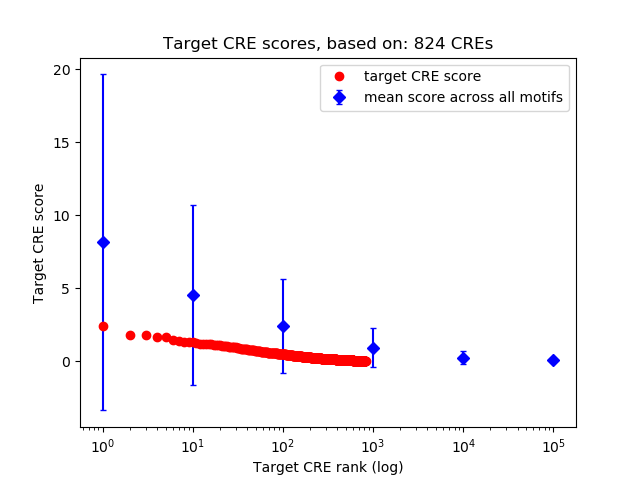
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_32684554_32685266 | 2.38 |
3425401B19Rik |
RIKEN cDNA 3425401B19 gene |
362 |
0.71 |
| chr9_58156326_58158409 | 1.83 |
Islr |
immunoglobulin superfamily containing leucine-rich repeat |
1178 |
0.35 |
| chr10_45337719_45337889 | 1.79 |
Bves |
blood vessel epicardial substance |
2032 |
0.3 |
| chr7_112660648_112660799 | 1.70 |
Gm45473 |
predicted gene 45473 |
7941 |
0.16 |
| chr10_5287995_5289689 | 1.68 |
Gm23573 |
predicted gene, 23573 |
68331 |
0.12 |
| chr2_139679952_139680200 | 1.44 |
Ism1 |
isthmin 1, angiogenesis inhibitor |
1898 |
0.34 |
| chr9_99709458_99709950 | 1.41 |
Cldn18 |
claudin 18 |
305 |
0.84 |
| chr3_145991092_145991594 | 1.34 |
Syde2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
2328 |
0.28 |
| chr17_74903463_74903703 | 1.32 |
Ttc27 |
tetratricopeptide repeat domain 27 |
41662 |
0.12 |
| chr1_133245733_133247220 | 1.31 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
370 |
0.84 |
| chr2_180388160_180389480 | 1.25 |
Mir1a-1 |
microRNA 1a-1 |
228 |
0.89 |
| chr2_7079530_7079727 | 1.20 |
Celf2 |
CUGBP, Elav-like family member 2 |
1579 |
0.52 |
| chr9_14612272_14612966 | 1.20 |
Amotl1 |
angiomotin-like 1 |
1967 |
0.22 |
| chr16_95703800_95705386 | 1.18 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
1746 |
0.39 |
| chrX_157699166_157699560 | 1.18 |
Smpx |
small muscle protein, X-linked |
103 |
0.96 |
| chr16_42907917_42908393 | 1.17 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
503 |
0.78 |
| chr14_101841178_101841822 | 1.14 |
Lmo7 |
LIM domain only 7 |
681 |
0.79 |
| chr8_126978786_126979425 | 1.10 |
Gm38574 |
predicted gene, 38574 |
3127 |
0.19 |
| chr15_57761421_57761679 | 1.09 |
9330154K18Rik |
RIKEN cDNA 9330154K18 gene |
22884 |
0.19 |
| chr2_92140355_92141595 | 1.09 |
Phf21a |
PHD finger protein 21A |
43131 |
0.11 |
| chr2_74681727_74681878 | 1.08 |
Hoxd11 |
homeobox D11 |
521 |
0.45 |
| chr4_133555894_133556045 | 1.07 |
Nr0b2 |
nuclear receptor subfamily 0, group B, member 2 |
2593 |
0.14 |
| chr6_49096738_49097482 | 1.03 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
2899 |
0.16 |
| chr7_45019658_45019809 | 1.03 |
Rras |
related RAS viral (r-ras) oncogene |
263 |
0.74 |
| chr6_52174920_52176658 | 1.01 |
Hoxaas3 |
Hoxa cluster antisense RNA 3 |
554 |
0.41 |
| chr6_34599135_34600037 | 0.98 |
Cald1 |
caldesmon 1 |
966 |
0.58 |
| chr10_126981839_126982215 | 0.98 |
Ctdsp2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
2998 |
0.11 |
| chr8_25225225_25225470 | 0.97 |
Tacc1 |
transforming, acidic coiled-coil containing protein 1 |
15516 |
0.19 |
| chr9_115356841_115357705 | 0.96 |
Mir467h |
microRNA 467h |
24546 |
0.12 |
| chr7_16892200_16892882 | 0.95 |
Gng8 |
guanine nucleotide binding protein (G protein), gamma 8 |
220 |
0.83 |
| chr7_100926314_100930096 | 0.93 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
3902 |
0.17 |
| chr17_12766020_12767485 | 0.92 |
Igf2r |
insulin-like growth factor 2 receptor |
2912 |
0.17 |
| chr6_97146396_97147185 | 0.88 |
Eogt |
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
994 |
0.39 |
| chr15_3581853_3582700 | 0.87 |
Ghr |
growth hormone receptor |
320 |
0.92 |
| chr13_18790213_18790483 | 0.86 |
Vps41 |
VPS41 HOPS complex subunit |
10505 |
0.27 |
| chr10_126391702_126391947 | 0.84 |
1700025K04Rik |
RIKEN cDNA 1700025K04 gene |
19849 |
0.25 |
| chr12_40016524_40017443 | 0.84 |
Arl4a |
ADP-ribosylation factor-like 4A |
2499 |
0.29 |
| chr13_112465692_112466190 | 0.84 |
Il6st |
interleukin 6 signal transducer |
1652 |
0.33 |
| chr16_95453226_95453447 | 0.81 |
Erg |
ETS transcription factor |
5909 |
0.29 |
| chr4_8236769_8237012 | 0.81 |
Gm25355 |
predicted gene, 25355 |
1308 |
0.43 |
| chr11_100931288_100932301 | 0.80 |
Stat3 |
signal transducer and activator of transcription 3 |
7586 |
0.15 |
| chr11_45175304_45175455 | 0.79 |
Gm12160 |
predicted gene 12160 |
14436 |
0.19 |
| chr14_54575380_54575961 | 0.77 |
Ajuba |
ajuba LIM protein |
1888 |
0.16 |
| chr4_46664011_46665033 | 0.76 |
Tbc1d2 |
TBC1 domain family, member 2 |
14313 |
0.19 |
| chr17_75440969_75441455 | 0.76 |
Rasgrp3 |
RAS, guanyl releasing protein 3 |
5286 |
0.29 |
| chr3_107092040_107092191 | 0.76 |
Gm27008 |
predicted gene, 27008 |
313 |
0.81 |
| chr5_77126646_77126869 | 0.75 |
Gm15831 |
predicted gene 15831 |
4409 |
0.15 |
| chr11_43383698_43384165 | 0.74 |
Gm12148 |
predicted gene 12148 |
1551 |
0.33 |
| chr2_48380324_48380475 | 0.74 |
Gm25338 |
predicted gene, 25338 |
3939 |
0.26 |
| chr5_114457519_114457716 | 0.74 |
Mvk |
mevalonate kinase |
5197 |
0.16 |
| chr3_131156467_131156618 | 0.73 |
Gm42449 |
predicted gene 42449 |
19757 |
0.17 |
| chr1_135799428_135800706 | 0.72 |
Tnni1 |
troponin I, skeletal, slow 1 |
234 |
0.9 |
| chr16_88560099_88560462 | 0.72 |
Cldn8 |
claudin 8 |
2903 |
0.2 |
| chr1_187662217_187663036 | 0.72 |
Gm37929 |
predicted gene, 37929 |
735 |
0.68 |
| chr5_151107821_151109068 | 0.71 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
291 |
0.94 |
| chr15_84131300_84131654 | 0.69 |
Gm33432 |
predicted gene, 33432 |
4173 |
0.13 |
| chr4_152115684_152116767 | 0.67 |
Tnfrsf25 |
tumor necrosis factor receptor superfamily, member 25 |
118 |
0.94 |
| chr3_137341050_137342448 | 0.66 |
Emcn |
endomucin |
575 |
0.82 |
| chr1_164461051_164462265 | 0.66 |
Gm32391 |
predicted gene, 32391 |
784 |
0.55 |
| chr2_9906124_9906915 | 0.65 |
Gm13262 |
predicted gene 13262 |
341 |
0.81 |
| chr15_91703120_91703338 | 0.64 |
Lrrk2 |
leucine-rich repeat kinase 2 |
30054 |
0.19 |
| chr7_90231440_90231794 | 0.63 |
Gm45220 |
predicted gene 45220 |
30339 |
0.11 |
| chr9_44498132_44499953 | 0.63 |
Bcl9l |
B cell CLL/lymphoma 9-like |
94 |
0.91 |
| chr15_98603382_98604193 | 0.63 |
Adcy6 |
adenylate cyclase 6 |
944 |
0.36 |
| chr11_72408624_72409355 | 0.63 |
Smtnl2 |
smoothelin-like 2 |
2722 |
0.18 |
| chr18_35915478_35915629 | 0.62 |
C330008A17Rik |
RIKEN cDNA C330008A17 gene |
2421 |
0.18 |
| chr1_12718823_12719219 | 0.61 |
Sulf1 |
sulfatase 1 |
443 |
0.85 |
| chr2_163377082_163377233 | 0.60 |
Jph2 |
junctophilin 2 |
20792 |
0.13 |
| chr2_91125839_91126091 | 0.59 |
Mybpc3 |
myosin binding protein C, cardiac |
7821 |
0.12 |
| chr11_8486434_8486685 | 0.59 |
Tns3 |
tensin 3 |
17884 |
0.28 |
| chr2_155313780_155313931 | 0.58 |
Pigu |
phosphatidylinositol glycan anchor biosynthesis, class U |
9163 |
0.16 |
| chr3_93522538_93523109 | 0.58 |
S100a11 |
S100 calcium binding protein A11 |
2335 |
0.17 |
| chr6_30741945_30743001 | 0.58 |
Mest |
mesoderm specific transcript |
384 |
0.72 |
| chr18_61665808_61666455 | 0.58 |
Carmn |
cardiac mesoderm enhancer-associated non-coding RNA |
594 |
0.59 |
| chr6_99469204_99469432 | 0.57 |
Gm22328 |
predicted gene, 22328 |
15384 |
0.18 |
| chr3_115712293_115713025 | 0.57 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
2413 |
0.28 |
| chr11_76381220_76381371 | 0.57 |
Nxn |
nucleoredoxin |
17773 |
0.17 |
| chr8_126867391_126867646 | 0.56 |
Gm31718 |
predicted gene, 31718 |
8920 |
0.2 |
| chr19_57239099_57239543 | 0.56 |
Ablim1 |
actin-binding LIM protein 1 |
8 |
0.98 |
| chr11_55417729_55419811 | 0.55 |
Sparc |
secreted acidic cysteine rich glycoprotein |
1128 |
0.44 |
| chr5_120777078_120777234 | 0.55 |
Oas3 |
2'-5' oligoadenylate synthetase 3 |
484 |
0.47 |
| chr9_43231400_43231766 | 0.54 |
Oaf |
out at first homolog |
6491 |
0.16 |
| chr1_160833263_160833414 | 0.54 |
Gm38060 |
predicted gene, 38060 |
5830 |
0.13 |
| chr13_52100486_52100637 | 0.54 |
4921525O09Rik |
RIKEN cDNA 4921525O09 gene |
2019 |
0.33 |
| chr6_70723615_70723766 | 0.54 |
Igkj5 |
immunoglobulin kappa joining 5 |
196 |
0.69 |
| chr18_53469057_53469955 | 0.54 |
Prdm6 |
PR domain containing 6 |
3475 |
0.33 |
| chr6_20264570_20264721 | 0.53 |
Gm42581 |
predicted gene 42581 |
476592 |
0.01 |
| chr2_114052280_114052567 | 0.52 |
Actc1 |
actin, alpha, cardiac muscle 1 |
464 |
0.75 |
| chr19_6854909_6856205 | 0.52 |
Ccdc88b |
coiled-coil domain containing 88B |
319 |
0.8 |
| chr3_9827707_9828674 | 0.51 |
Gm24786 |
predicted gene, 24786 |
1538 |
0.39 |
| chr18_53462509_53463403 | 0.51 |
Prdm6 |
PR domain containing 6 |
627 |
0.8 |
| chr8_76710038_76710189 | 0.51 |
Gm27355 |
predicted gene, 27355 |
90831 |
0.09 |
| chr14_120479808_120480690 | 0.50 |
Rap2a |
RAS related protein 2a |
1805 |
0.45 |
| chr15_102752755_102752906 | 0.50 |
Calcoco1 |
calcium binding and coiled coil domain 1 |
30652 |
0.11 |
| chr17_78934273_78934424 | 0.50 |
Cebpz |
CCAAT/enhancer binding protein zeta |
2689 |
0.17 |
| chr5_24928061_24928212 | 0.50 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
19159 |
0.15 |
| chr2_147690687_147690838 | 0.50 |
A530006G24Rik |
RIKEN cDNA A530006G24 gene |
19931 |
0.19 |
| chr12_57547376_57547527 | 0.49 |
Foxa1 |
forkhead box A1 |
535 |
0.73 |
| chr8_22506948_22507557 | 0.49 |
Slc20a2 |
solute carrier family 20, member 2 |
1026 |
0.46 |
| chr5_148341058_148341274 | 0.49 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
1563 |
0.45 |
| chr2_148036751_148037106 | 0.48 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
1342 |
0.4 |
| chr4_108838068_108838658 | 0.48 |
Txndc12 |
thioredoxin domain containing 12 (endoplasmic reticulum) |
3728 |
0.16 |
| chr17_29030374_29030534 | 0.48 |
Gm16196 |
predicted gene 16196 |
2114 |
0.14 |
| chr16_38709025_38709629 | 0.47 |
Arhgap31 |
Rho GTPase activating protein 31 |
3571 |
0.19 |
| chr10_110921543_110922443 | 0.47 |
Csrp2 |
cysteine and glycine-rich protein 2 |
1788 |
0.28 |
| chr2_180692538_180693057 | 0.47 |
Gm22502 |
predicted gene, 22502 |
5266 |
0.13 |
| chr11_96316025_96316970 | 0.47 |
Mir10a |
microRNA 10a |
668 |
0.4 |
| chr2_180389757_180390438 | 0.46 |
Mir1a-1 |
microRNA 1a-1 |
1049 |
0.41 |
| chr19_42936859_42937183 | 0.46 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
157043 |
0.03 |
| chr12_59095360_59096062 | 0.46 |
Mia2 |
MIA SH3 domain ER export factor 2 |
88 |
0.93 |
| chr9_118082500_118083548 | 0.46 |
Azi2 |
5-azacytidine induced gene 2 |
23968 |
0.18 |
| chr11_20914754_20915017 | 0.46 |
Gm23681 |
predicted gene, 23681 |
22816 |
0.18 |
| chr2_122298256_122299570 | 0.46 |
Duoxa2 |
dual oxidase maturation factor 2 |
13 |
0.88 |
| chr11_30261256_30261407 | 0.45 |
Sptbn1 |
spectrin beta, non-erythrocytic 1 |
5883 |
0.23 |
| chr11_19012477_19012923 | 0.45 |
Meis1 |
Meis homeobox 1 |
698 |
0.49 |
| chr17_47989101_47989367 | 0.45 |
Gm14871 |
predicted gene 14871 |
14338 |
0.14 |
| chrX_7673653_7673804 | 0.45 |
Plp2 |
proteolipid protein 2 |
2338 |
0.12 |
| chr13_47148646_47148901 | 0.43 |
A930002C04Rik |
RIKEN cDNA A930002C04 gene |
103 |
0.95 |
| chr5_140890731_140891173 | 0.43 |
Card11 |
caspase recruitment domain family, member 11 |
8747 |
0.24 |
| chr13_112651975_112652409 | 0.42 |
Ddx4 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
109 |
0.96 |
| chr11_89544915_89545658 | 0.42 |
Ankfn1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
6730 |
0.32 |
| chr4_29092846_29093062 | 0.41 |
Gm11916 |
predicted gene 11916 |
2349 |
0.32 |
| chr11_68956320_68957720 | 0.41 |
Arhgef15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
460 |
0.64 |
| chr7_131032506_131032724 | 0.41 |
Dmbt1 |
deleted in malignant brain tumors 1 |
546 |
0.79 |
| chr16_96362648_96363004 | 0.41 |
Igsf5 |
immunoglobulin superfamily, member 5 |
1032 |
0.37 |
| chr2_32311532_32312331 | 0.40 |
Dnm1 |
dynamin 1 |
958 |
0.33 |
| chr3_144617679_144618017 | 0.40 |
Rpl9-ps8 |
ribosomal protein L9, pseudogene 8 |
8116 |
0.15 |
| chr17_29368961_29369571 | 0.40 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
287 |
0.85 |
| chr3_153906931_153907082 | 0.39 |
Msh4 |
mutS homolog 4 |
868 |
0.37 |
| chr8_123157519_123158497 | 0.39 |
Sult5a1 |
sulfotransferase family 5A, member 1 |
220 |
0.84 |
| chr9_123844355_123844506 | 0.39 |
Fyco1 |
FYVE and coiled-coil domain containing 1 |
176 |
0.93 |
| chr1_14756748_14757310 | 0.39 |
Msc |
musculin |
1037 |
0.45 |
| chr17_25102221_25102372 | 0.38 |
Telo2 |
telomere maintenance 2 |
870 |
0.42 |
| chr2_32318746_32319267 | 0.38 |
Mir199b |
microRNA 199b |
546 |
0.42 |
| chr17_56604701_56604852 | 0.38 |
Safb |
scaffold attachment factor B |
417 |
0.71 |
| chr3_102085891_102086191 | 0.37 |
Casq2 |
calsequestrin 2 |
374 |
0.82 |
| chr3_79229375_79229526 | 0.37 |
4921511C10Rik |
RIKEN cDNA 4921511C10 gene |
16531 |
0.18 |
| chr7_19824650_19824851 | 0.37 |
Bcl3 |
B cell leukemia/lymphoma 3 |
1980 |
0.14 |
| chr6_52154157_52154308 | 0.37 |
Hotairm1 |
Hoxa transcript antisense RNA, myeloid-specific 1 |
2670 |
0.09 |
| chr12_117743663_117743926 | 0.37 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
3199 |
0.27 |
| chr5_119829778_119830485 | 0.36 |
Gm43050 |
predicted gene 43050 |
1660 |
0.31 |
| chr11_115133328_115133870 | 0.35 |
Cd300lf |
CD300 molecule like family member F |
392 |
0.78 |
| chr12_70827053_70827698 | 0.35 |
Frmd6 |
FERM domain containing 6 |
1686 |
0.31 |
| chr18_37750859_37751188 | 0.35 |
Pcdhgb7 |
protocadherin gamma subfamily B, 7 |
577 |
0.38 |
| chr9_22277194_22277360 | 0.35 |
Zfp810 |
zinc finger protein 810 |
6626 |
0.09 |
| chr15_102998770_103001153 | 0.35 |
Hoxc6 |
homeobox C6 |
568 |
0.54 |
| chr5_41758847_41759075 | 0.35 |
Nkx3-2 |
NK3 homeobox 2 |
5540 |
0.25 |
| chr12_69296651_69297715 | 0.34 |
Klhdc2 |
kelch domain containing 2 |
428 |
0.72 |
| chr4_130802234_130802571 | 0.34 |
Sdc3 |
syndecan 3 |
9536 |
0.11 |
| chr18_44557958_44558665 | 0.34 |
Mcc |
mutated in colorectal cancers |
38795 |
0.19 |
| chr1_120176211_120176597 | 0.34 |
3110009E18Rik |
RIKEN cDNA 3110009E18 gene |
15924 |
0.2 |
| chr10_122010922_122011073 | 0.34 |
Srgap1 |
SLIT-ROBO Rho GTPase activating protein 1 |
36311 |
0.13 |
| chr8_117493765_117493916 | 0.33 |
Plcg2 |
phospholipase C, gamma 2 |
4451 |
0.21 |
| chr4_155839735_155840422 | 0.33 |
Mxra8os |
matrix-remodelling associated 8, opposite strand |
100 |
0.67 |
| chr13_41251840_41252333 | 0.33 |
Smim13 |
small integral membrane protein 13 |
2242 |
0.2 |
| chr10_53343547_53344097 | 0.33 |
Gm47644 |
predicted gene, 47644 |
412 |
0.77 |
| chr1_171501506_171502096 | 0.33 |
Gm24871 |
predicted gene, 24871 |
1362 |
0.2 |
| chr13_32493561_32493930 | 0.33 |
Gm48073 |
predicted gene, 48073 |
42670 |
0.17 |
| chr13_9128006_9128583 | 0.33 |
Larp4b |
La ribonucleoprotein domain family, member 4B |
7921 |
0.16 |
| chr5_34217945_34218096 | 0.33 |
Gm42848 |
predicted gene 42848 |
3142 |
0.14 |
| chr14_6637179_6637341 | 0.33 |
Gm3460 |
predicted gene 3460 |
14842 |
0.14 |
| chr4_82930111_82930271 | 0.33 |
Frem1 |
Fras1 related extracellular matrix protein 1 |
5399 |
0.25 |
| chr19_43609438_43610955 | 0.32 |
Gm20467 |
predicted gene 20467 |
429 |
0.71 |
| chr10_84438288_84438980 | 0.32 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
1963 |
0.25 |
| chr10_25449303_25449917 | 0.32 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
7999 |
0.21 |
| chr19_38054245_38054734 | 0.32 |
I830134H01Rik |
RIKEN cDNA I830134H01 gene |
39 |
0.9 |
| chr6_136873814_136874977 | 0.32 |
Mgp |
matrix Gla protein |
1386 |
0.26 |
| chr1_19223778_19223929 | 0.32 |
Tfap2b |
transcription factor AP-2 beta |
9974 |
0.19 |
| chr16_91348159_91348310 | 0.30 |
Gm24695 |
predicted gene, 24695 |
1550 |
0.28 |
| chr17_56591227_56591378 | 0.30 |
Safb |
scaffold attachment factor B |
2462 |
0.16 |
| chr15_103036300_103036759 | 0.30 |
Hoxc4 |
homeobox C4 |
2134 |
0.16 |
| chr10_110927145_110928031 | 0.30 |
Csrp2 |
cysteine and glycine-rich protein 2 |
3613 |
0.19 |
| chr12_16739495_16739811 | 0.30 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
475 |
0.79 |
| chr6_116651389_116652299 | 0.30 |
Depp1 |
DEPP1 autophagy regulator |
1148 |
0.32 |
| chr16_37882910_37883557 | 0.30 |
Gm15725 |
predicted gene 15725 |
691 |
0.61 |
| chr2_150650997_150651161 | 0.30 |
Acss1 |
acyl-CoA synthetase short-chain family member 1 |
12964 |
0.12 |
| chr10_79806462_79808470 | 0.30 |
Palm |
paralemmin |
785 |
0.35 |
| chr11_115416733_115417187 | 0.30 |
Atp5h |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
58 |
0.93 |
| chr11_88275982_88276133 | 0.29 |
Ccdc182 |
coiled-coil domain containing 182 |
18001 |
0.15 |
| chr8_117692115_117692317 | 0.29 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
9688 |
0.13 |
| chr16_90405483_90405634 | 0.29 |
Hunk |
hormonally upregulated Neu-associated kinase |
5014 |
0.2 |
| chr9_98968051_98968202 | 0.29 |
Foxl2os |
forkhead box L2, opposite strand |
12827 |
0.11 |
| chr17_28272737_28272895 | 0.29 |
Ppard |
peroxisome proliferator activator receptor delta |
697 |
0.55 |
| chr17_12689035_12689208 | 0.29 |
Igf2r |
insulin-like growth factor 2 receptor |
12158 |
0.15 |
| chr3_127779635_127780565 | 0.28 |
Alpk1 |
alpha-kinase 1 |
390 |
0.77 |
| chr8_62284047_62284979 | 0.28 |
Gm2961 |
predicted gene 2961 |
90301 |
0.09 |
| chr12_112490769_112491162 | 0.28 |
A530016L24Rik |
RIKEN cDNA A530016L24 gene |
1491 |
0.35 |
| chr13_56609245_56610611 | 0.28 |
Tgfbi |
transforming growth factor, beta induced |
342 |
0.88 |
| chr8_104108186_104108431 | 0.28 |
Gm29682 |
predicted gene, 29682 |
2435 |
0.2 |
| chr16_19981596_19982182 | 0.27 |
Klhl6 |
kelch-like 6 |
1041 |
0.58 |
| chr11_116821547_116822325 | 0.27 |
Mxra7 |
matrix-remodelling associated 7 |
6067 |
0.12 |
| chrX_7316516_7317667 | 0.27 |
Clcn5 |
chloride channel, voltage-sensitive 5 |
2079 |
0.26 |
| chr4_129575140_129575515 | 0.27 |
Lck |
lymphocyte protein tyrosine kinase |
1686 |
0.17 |
| chr6_92071359_92071510 | 0.27 |
Fgd5 |
FYVE, RhoGEF and PH domain containing 5 |
246 |
0.91 |
| chr13_58122870_58123670 | 0.27 |
Hnrnpa0 |
heterogeneous nuclear ribonucleoprotein A0 |
5286 |
0.14 |
| chr10_22821973_22822241 | 0.27 |
Tcf21 |
transcription factor 21 |
1933 |
0.29 |
| chr4_114422836_114422987 | 0.27 |
Trabd2b |
TraB domain containing 2B |
16187 |
0.28 |
| chr4_8963467_8963668 | 0.26 |
Rps18-ps2 |
ribosomal protein S18, pseudogene 2 |
22308 |
0.26 |
| chr7_140954618_140955957 | 0.26 |
Gm45717 |
predicted gene 45717 |
650 |
0.32 |
| chr15_6861574_6861725 | 0.26 |
Osmr |
oncostatin M receptor |
12608 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.2 | 0.9 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.2 | 0.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.2 | 0.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.7 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.1 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.9 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.2 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.1 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.3 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0071864 | positive regulation of cell proliferation in bone marrow(GO:0071864) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:1904683 | regulation of metalloendopeptidase activity(GO:1904683) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.0 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.5 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:0072162 | paramesonephric duct development(GO:0061205) metanephric mesenchymal cell differentiation(GO:0072162) |
| 0.0 | 0.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.4 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.0 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0060406 | regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.0 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.0 | GO:0030730 | sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.2 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |