Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
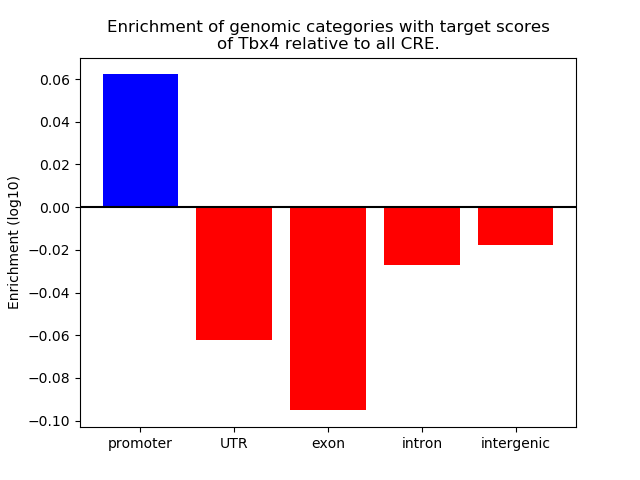
Results for Tbx4
Z-value: 0.22
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSMUSG00000000094.6 | T-box 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_85894577_85895744 | Tbx4 | 1605 | 0.358953 | 0.61 | 2.3e-07 | Click! |
| chr11_85894276_85894531 | Tbx4 | 2362 | 0.268991 | 0.50 | 4.0e-05 | Click! |
| chr11_85888239_85889268 | Tbx4 | 1310 | 0.413354 | 0.46 | 2.1e-04 | Click! |
| chr11_85889787_85891444 | Tbx4 | 552 | 0.745685 | 0.43 | 5.2e-04 | Click! |
| chr11_85892296_85892447 | Tbx4 | 2308 | 0.270450 | 0.43 | 6.5e-04 | Click! |
Activity of the Tbx4 motif across conditions
Conditions sorted by the z-value of the Tbx4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
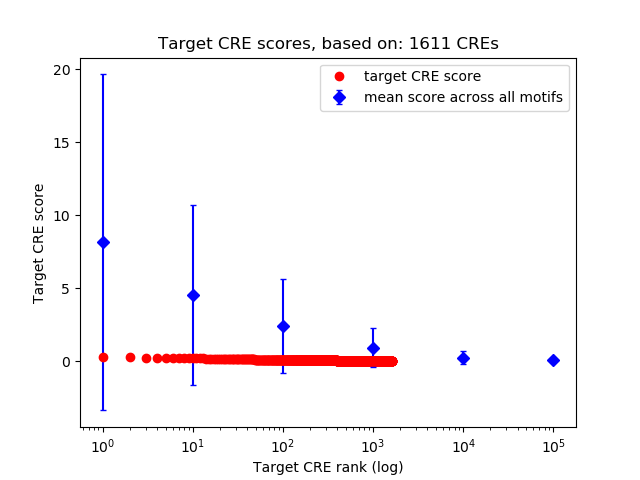
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_163395773_163396078 | 0.29 |
Jph2 |
junctophilin 2 |
2024 |
0.24 |
| chr2_163396102_163396417 | 0.27 |
Jph2 |
junctophilin 2 |
1690 |
0.28 |
| chr3_30546274_30547260 | 0.25 |
Mecom |
MDS1 and EVI1 complex locus |
1241 |
0.35 |
| chr9_119962629_119963694 | 0.25 |
Ttc21a |
tetratricopeptide repeat domain 21A |
1543 |
0.24 |
| chr1_34116331_34116653 | 0.25 |
Mir6896 |
microRNA 6896 |
867 |
0.57 |
| chr3_19646421_19646748 | 0.22 |
Trim55 |
tripartite motif-containing 55 |
2076 |
0.26 |
| chr9_99709458_99709950 | 0.21 |
Cldn18 |
claudin 18 |
305 |
0.84 |
| chr1_51291546_51292409 | 0.20 |
Cavin2 |
caveolae associated 2 |
2851 |
0.27 |
| chr13_63241575_63242090 | 0.20 |
Aopep |
aminopeptidase O |
1724 |
0.2 |
| chr16_44015370_44016774 | 0.20 |
Gramd1c |
GRAM domain containing 1C |
364 |
0.83 |
| chr12_103336858_103338251 | 0.20 |
Gm15523 |
predicted gene 15523 |
648 |
0.43 |
| chr8_57324709_57326732 | 0.19 |
Hand2os1 |
Hand2, opposite strand 1 |
1487 |
0.3 |
| chr16_36989294_36990265 | 0.19 |
Fbxo40 |
F-box protein 40 |
688 |
0.6 |
| chr8_25027794_25028434 | 0.19 |
Gm16933 |
predicted gene, 16933 |
8938 |
0.12 |
| chr2_91120847_91121416 | 0.19 |
Mybpc3 |
myosin binding protein C, cardiac |
2987 |
0.16 |
| chr3_155054087_155054557 | 0.19 |
Tnni3k |
TNNI3 interacting kinase |
1028 |
0.55 |
| chr11_74526613_74527288 | 0.18 |
Rap1gap2 |
RAP1 GTPase activating protein 2 |
4199 |
0.25 |
| chr17_81737002_81738450 | 0.18 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
651 |
0.81 |
| chr8_61901105_61901625 | 0.17 |
Palld |
palladin, cytoskeletal associated protein |
1304 |
0.42 |
| chr9_44069968_44070511 | 0.17 |
Usp2 |
ubiquitin specific peptidase 2 |
796 |
0.33 |
| chr11_95841518_95842457 | 0.16 |
Abi3 |
ABI gene family, member 3 |
111 |
0.73 |
| chr8_27261786_27261937 | 0.16 |
Eif4ebp1 |
eukaryotic translation initiation factor 4E binding protein 1 |
1532 |
0.26 |
| chr1_98132044_98132246 | 0.16 |
1810006J02Rik |
RIKEN cDNA 1810006J02 gene |
678 |
0.7 |
| chr2_26585065_26586013 | 0.16 |
Egfl7 |
EGF-like domain 7 |
1091 |
0.24 |
| chr11_99042414_99042858 | 0.15 |
Igfbp4 |
insulin-like growth factor binding protein 4 |
450 |
0.73 |
| chr13_46423537_46423785 | 0.15 |
Rbm24 |
RNA binding motif protein 24 |
1839 |
0.41 |
| chr9_45934543_45935100 | 0.15 |
Tagln |
transgelin |
753 |
0.43 |
| chr6_112728907_112729181 | 0.15 |
Rad18 |
RAD18 E3 ubiquitin protein ligase |
32358 |
0.16 |
| chr6_4005591_4005747 | 0.14 |
Gng11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1765 |
0.31 |
| chr16_95703800_95705386 | 0.14 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
1746 |
0.39 |
| chr2_92140355_92141595 | 0.14 |
Phf21a |
PHD finger protein 21A |
43131 |
0.11 |
| chr15_3580312_3581004 | 0.14 |
Ghr |
growth hormone receptor |
1184 |
0.57 |
| chr2_32318746_32319267 | 0.14 |
Mir199b |
microRNA 199b |
546 |
0.42 |
| chr18_35120289_35121263 | 0.14 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
1334 |
0.45 |
| chr3_138067510_138069146 | 0.13 |
Gm5105 |
predicted gene 5105 |
940 |
0.46 |
| chr9_123480978_123481680 | 0.13 |
Limd1 |
LIM domains containing 1 |
719 |
0.66 |
| chr5_134745620_134746234 | 0.13 |
Gm30003 |
predicted gene, 30003 |
1115 |
0.35 |
| chr5_147723576_147724353 | 0.13 |
Flt1 |
FMS-like tyrosine kinase 1 |
2024 |
0.35 |
| chr12_84643018_84643308 | 0.13 |
Vrtn |
vertebrae development associated |
267 |
0.89 |
| chr7_140954618_140955957 | 0.13 |
Gm45717 |
predicted gene 45717 |
650 |
0.32 |
| chr15_12010211_12011091 | 0.13 |
Sub1 |
SUB1 homolog, transcriptional regulator |
13668 |
0.16 |
| chr2_145787122_145787273 | 0.13 |
Rin2 |
Ras and Rab interactor 2 |
1035 |
0.61 |
| chr1_153329741_153330128 | 0.13 |
Lamc1 |
laminin, gamma 1 |
2852 |
0.24 |
| chr13_23913630_23913830 | 0.12 |
Slc17a4 |
solute carrier family 17 (sodium phosphate), member 4 |
1238 |
0.27 |
| chr8_57340613_57341952 | 0.12 |
5033428I22Rik |
RIKEN cDNA 5033428I22 gene |
482 |
0.73 |
| chr19_53557067_53557881 | 0.12 |
Gm50394 |
predicted gene, 50394 |
27819 |
0.11 |
| chr16_87553258_87554074 | 0.12 |
Map3k7cl |
Map3k7 C-terminal like |
336 |
0.84 |
| chr2_35627849_35628114 | 0.12 |
Dab2ip |
disabled 2 interacting protein |
5821 |
0.24 |
| chr10_22813360_22814229 | 0.12 |
Gm10824 |
predicted gene 10824 |
1955 |
0.28 |
| chr5_75152837_75154692 | 0.12 |
Gm42802 |
predicted gene 42802 |
111 |
0.58 |
| chr10_7831857_7832952 | 0.12 |
Zc3h12d |
zinc finger CCCH type containing 12D |
66 |
0.95 |
| chr10_120900758_120900909 | 0.12 |
Gm4473 |
predicted gene 4473 |
1676 |
0.25 |
| chr14_62798730_62799095 | 0.12 |
Gm18146 |
predicted gene, 18146 |
19215 |
0.12 |
| chr8_104101710_104103631 | 0.12 |
Cdh5 |
cadherin 5 |
1045 |
0.43 |
| chr11_54962647_54963034 | 0.12 |
Tnip1 |
TNFAIP3 interacting protein 1 |
69 |
0.97 |
| chr10_127749619_127751660 | 0.12 |
Gpr182 |
G protein-coupled receptor 182 |
1093 |
0.28 |
| chr7_130576739_130577783 | 0.12 |
Tacc2 |
transforming, acidic coiled-coil containing protein 2 |
177 |
0.94 |
| chr3_60504423_60505028 | 0.12 |
Mbnl1 |
muscleblind like splicing factor 1 |
3447 |
0.29 |
| chr2_181241290_181242534 | 0.11 |
Helz2 |
helicase with zinc finger 2, transcriptional coactivator |
51 |
0.95 |
| chr14_26439572_26439816 | 0.11 |
Slmap |
sarcolemma associated protein |
473 |
0.78 |
| chr14_101510884_101511350 | 0.11 |
Tbc1d4 |
TBC1 domain family, member 4 |
3791 |
0.3 |
| chr12_82122852_82123243 | 0.11 |
n-R5s62 |
nuclear encoded rRNA 5S 62 |
27421 |
0.18 |
| chr1_74748322_74749490 | 0.11 |
Prkag3 |
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
2 |
0.96 |
| chr15_101411658_101413570 | 0.11 |
Krt7 |
keratin 7 |
165 |
0.64 |
| chr4_141302549_141302700 | 0.11 |
Epha2 |
Eph receptor A2 |
217 |
0.88 |
| chr9_44087058_44087369 | 0.11 |
Usp2 |
ubiquitin specific peptidase 2 |
23 |
0.93 |
| chr13_41345830_41346374 | 0.11 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
13145 |
0.14 |
| chr5_32990256_32991187 | 0.11 |
Gm43693 |
predicted gene 43693 |
6151 |
0.16 |
| chr13_102036243_102036435 | 0.11 |
Gm38133 |
predicted gene, 38133 |
79696 |
0.1 |
| chr12_117200442_117200898 | 0.11 |
Gm10421 |
predicted gene 10421 |
49019 |
0.16 |
| chr8_41040672_41042201 | 0.11 |
Mtus1 |
mitochondrial tumor suppressor 1 |
430 |
0.76 |
| chr15_98605967_98606147 | 0.11 |
Adcy6 |
adenylate cyclase 6 |
1231 |
0.27 |
| chr14_49244860_49245366 | 0.11 |
ccdc198 |
coiled-coil domain containing 198 |
284 |
0.89 |
| chr7_16892200_16892882 | 0.11 |
Gng8 |
guanine nucleotide binding protein (G protein), gamma 8 |
220 |
0.83 |
| chr9_55283384_55284127 | 0.11 |
Nrg4 |
neuregulin 4 |
130 |
0.96 |
| chr14_49794107_49794296 | 0.11 |
Gm22989 |
predicted gene, 22989 |
5917 |
0.14 |
| chr6_115984719_115988278 | 0.11 |
Plxnd1 |
plexin D1 |
8507 |
0.15 |
| chr16_58685640_58686063 | 0.11 |
Cpox |
coproporphyrinogen oxidase |
7995 |
0.13 |
| chr8_33916300_33917561 | 0.11 |
Rbpms |
RNA binding protein gene with multiple splicing |
12346 |
0.17 |
| chr19_56120628_56120882 | 0.11 |
Gm31912 |
predicted gene, 31912 |
14445 |
0.24 |
| chr6_51173127_51174365 | 0.10 |
Mir148a |
microRNA 148a |
96164 |
0.07 |
| chr2_144676648_144676927 | 0.10 |
Dtd1 |
D-tyrosyl-tRNA deacylase 1 |
1311 |
0.47 |
| chr11_63834406_63834560 | 0.10 |
Hmgb1-ps3 |
high mobility group box 1, pseudogene 3 |
12335 |
0.21 |
| chr3_116127845_116129091 | 0.10 |
Gm43109 |
predicted gene 43109 |
17 |
0.88 |
| chrX_159945522_159946439 | 0.10 |
Sh3kbp1 |
SH3-domain kinase binding protein 1 |
2962 |
0.3 |
| chr13_46424277_46424435 | 0.10 |
Rbm24 |
RNA binding motif protein 24 |
2534 |
0.34 |
| chr2_27079055_27080577 | 0.10 |
Adamtsl2 |
ADAMTS-like 2 |
437 |
0.73 |
| chr5_123318053_123318557 | 0.10 |
Gm15857 |
predicted gene 15857 |
7981 |
0.1 |
| chr3_50816196_50816734 | 0.10 |
Gm24503 |
predicted gene, 24503 |
22250 |
0.21 |
| chr3_49753928_49755610 | 0.10 |
Pcdh18 |
protocadherin 18 |
695 |
0.72 |
| chr5_137349923_137351747 | 0.10 |
Ephb4 |
Eph receptor B4 |
322 |
0.77 |
| chr10_18432775_18433290 | 0.10 |
Nhsl1 |
NHS-like 1 |
25357 |
0.21 |
| chr7_19011655_19012657 | 0.10 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
8112 |
0.07 |
| chr13_21996613_21997784 | 0.10 |
Prss16 |
protease, serine 16 (thymus) |
8024 |
0.06 |
| chr1_61850467_61850728 | 0.10 |
Pard3bos1 |
par-3 family cell polarity regulator beta, opposite strand 1 |
865 |
0.67 |
| chr3_131156467_131156618 | 0.10 |
Gm42449 |
predicted gene 42449 |
19757 |
0.17 |
| chr3_142474143_142474633 | 0.10 |
Gbp5 |
guanylate binding protein 5 |
19590 |
0.18 |
| chr10_62843668_62844437 | 0.10 |
Gm48353 |
predicted gene, 48353 |
15942 |
0.11 |
| chr3_94372701_94373986 | 0.10 |
Rorc |
RAR-related orphan receptor gamma |
549 |
0.5 |
| chr2_21034615_21035123 | 0.10 |
Gm13376 |
predicted gene 13376 |
62230 |
0.1 |
| chr5_101239366_101239752 | 0.10 |
Cycs-ps2 |
cytochrome c, pseudogene 2 |
66019 |
0.13 |
| chr9_32146234_32146589 | 0.10 |
Arhgap32 |
Rho GTPase activating protein 32 |
16841 |
0.17 |
| chr8_54571415_54571982 | 0.10 |
Asb5 |
ankyrin repeat and SOCs box-containing 5 |
272 |
0.9 |
| chr7_44478079_44478838 | 0.10 |
5430431A17Rik |
RIKEN cDNA 5430431A17 gene |
4920 |
0.07 |
| chr3_89832920_89834058 | 0.10 |
She |
src homology 2 domain-containing transforming protein E |
2119 |
0.21 |
| chr12_12904321_12905106 | 0.10 |
4930519A11Rik |
RIKEN cDNA 4930519A11 gene |
163 |
0.93 |
| chr2_166412306_166412566 | 0.10 |
Gm14268 |
predicted gene 14268 |
14249 |
0.21 |
| chr3_98280396_98280830 | 0.10 |
Hmgcs2 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
178 |
0.93 |
| chr6_124925245_124926316 | 0.10 |
Mlf2 |
myeloid leukemia factor 2 |
5606 |
0.08 |
| chr8_84021064_84022448 | 0.10 |
Palm3 |
paralemmin 3 |
285 |
0.75 |
| chr18_53466213_53466522 | 0.09 |
Prdm6 |
PR domain containing 6 |
1851 |
0.45 |
| chr4_117391533_117392402 | 0.09 |
Rnf220 |
ring finger protein 220 |
16510 |
0.16 |
| chr2_120268555_120268972 | 0.09 |
Pla2g4d |
phospholipase A2, group IVD |
20434 |
0.13 |
| chr13_113100448_113100599 | 0.09 |
Gzma |
granzyme A |
448 |
0.7 |
| chrX_152768117_152768457 | 0.09 |
Shroom2 |
shroom family member 2 |
1174 |
0.51 |
| chr13_51646613_51647088 | 0.09 |
Gm22806 |
predicted gene, 22806 |
1349 |
0.33 |
| chr19_6447935_6448212 | 0.09 |
Nrxn2 |
neurexin II |
1775 |
0.22 |
| chr17_86754057_86754608 | 0.09 |
Epas1 |
endothelial PAS domain protein 1 |
632 |
0.71 |
| chr5_100214874_100215472 | 0.09 |
2310034O05Rik |
RIKEN cDNA 2310034O05 gene |
4465 |
0.21 |
| chr10_76798931_76799264 | 0.09 |
Pcbp3 |
poly(rC) binding protein 3 |
1233 |
0.5 |
| chr17_69120419_69121537 | 0.09 |
Epb41l3 |
erythrocyte membrane protein band 4.1 like 3 |
6022 |
0.31 |
| chr8_3568500_3569092 | 0.09 |
Rps23rg1 |
ribosomal protein S23, retrogene 1 |
798 |
0.43 |
| chr1_189782721_189783385 | 0.09 |
Ptpn14 |
protein tyrosine phosphatase, non-receptor type 14 |
3583 |
0.24 |
| chr8_92265739_92266030 | 0.09 |
Gm45334 |
predicted gene 45334 |
32759 |
0.16 |
| chr5_128464543_128464766 | 0.09 |
Gm42500 |
predicted gene 42500 |
15750 |
0.15 |
| chr3_98245804_98245958 | 0.09 |
Gm42821 |
predicted gene 42821 |
2718 |
0.21 |
| chr5_140890731_140891173 | 0.09 |
Card11 |
caspase recruitment domain family, member 11 |
8747 |
0.24 |
| chr1_97975537_97975875 | 0.09 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
1362 |
0.45 |
| chr3_115712293_115713025 | 0.09 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
2413 |
0.28 |
| chr12_31270509_31272027 | 0.09 |
Lamb1 |
laminin B1 |
5388 |
0.14 |
| chr1_74303501_74304573 | 0.09 |
Tmbim1 |
transmembrane BAX inhibitor motif containing 1 |
83 |
0.93 |
| chr10_116750858_116751292 | 0.09 |
4930579P08Rik |
RIKEN cDNA 4930579P08 gene |
21822 |
0.16 |
| chr5_119742473_119742643 | 0.09 |
Tbx3os2 |
T-box 3, opposite strand 2 |
51340 |
0.1 |
| chr3_53165376_53165642 | 0.09 |
Gm43803 |
predicted gene 43803 |
52098 |
0.09 |
| chr4_133084556_133086193 | 0.09 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
19125 |
0.17 |
| chr5_29467659_29468760 | 0.09 |
Mnx1 |
motor neuron and pancreas homeobox 1 |
10261 |
0.16 |
| chr7_100657972_100658903 | 0.09 |
Plekhb1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
15 |
0.96 |
| chr11_78378001_78378152 | 0.09 |
Foxn1 |
forkhead box N1 |
8482 |
0.09 |
| chr9_44087472_44088439 | 0.09 |
Usp2 |
ubiquitin specific peptidase 2 |
765 |
0.37 |
| chr11_103096614_103097909 | 0.09 |
Plcd3 |
phospholipase C, delta 3 |
4330 |
0.13 |
| chr15_59030318_59030922 | 0.09 |
Mtss1 |
MTSS I-BAR domain containing 1 |
9976 |
0.21 |
| chr18_16436164_16436577 | 0.09 |
Gm7665 |
predicted pseudogene 7665 |
161426 |
0.04 |
| chr18_69118550_69118756 | 0.09 |
Gm23536 |
predicted gene, 23536 |
55467 |
0.12 |
| chr4_115088735_115089677 | 0.09 |
Pdzk1ip1 |
PDZK1 interacting protein 1 |
163 |
0.94 |
| chr5_101703476_101703804 | 0.09 |
9430085M18Rik |
RIKEN cDNA 9430085M18 gene |
54 |
0.87 |
| chr5_100844231_100844692 | 0.09 |
Gpat3 |
glycerol-3-phosphate acyltransferase 3 |
1252 |
0.27 |
| chr8_57335156_57335307 | 0.09 |
Gm34030 |
predicted gene, 34030 |
646 |
0.6 |
| chr10_115817324_115818606 | 0.09 |
Tspan8 |
tetraspanin 8 |
681 |
0.78 |
| chr7_102106752_102107605 | 0.09 |
Art5 |
ADP-ribosyltransferase 5 |
2311 |
0.16 |
| chr7_19295472_19296203 | 0.09 |
Rtn2 |
reticulon 2 (Z-band associated protein) |
4768 |
0.08 |
| chr8_70493071_70496051 | 0.09 |
Crlf1 |
cytokine receptor-like factor 1 |
1200 |
0.25 |
| chr5_120476252_120476535 | 0.09 |
Sds |
serine dehydratase |
133 |
0.92 |
| chr11_114478869_114479221 | 0.08 |
4932435O22Rik |
RIKEN cDNA 4932435O22 gene |
30185 |
0.2 |
| chr3_82141940_82142375 | 0.08 |
Gucy1a1 |
guanylate cyclase 1, soluble, alpha 1 |
2916 |
0.3 |
| chr2_32608081_32609404 | 0.08 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
25 |
0.94 |
| chr15_103383151_103383913 | 0.08 |
Gm37322 |
predicted gene, 37322 |
5461 |
0.13 |
| chr9_68234867_68235111 | 0.08 |
Gm28195 |
predicted gene 28195 |
40 |
0.98 |
| chr2_156717216_156718150 | 0.08 |
Dlgap4 |
DLG associated protein 4 |
2889 |
0.18 |
| chr16_91917498_91918270 | 0.08 |
Gm25245 |
predicted gene, 25245 |
3531 |
0.14 |
| chr6_34355142_34355293 | 0.08 |
Akr1b8 |
aldo-keto reductase family 1, member B8 |
898 |
0.49 |
| chr8_126587716_126588216 | 0.08 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
6020 |
0.24 |
| chr2_91125839_91126091 | 0.08 |
Mybpc3 |
myosin binding protein C, cardiac |
7821 |
0.12 |
| chrX_48400504_48400994 | 0.08 |
Bcorl1 |
BCL6 co-repressor-like 1 |
31481 |
0.15 |
| chr11_18873955_18876215 | 0.08 |
8430419K02Rik |
RIKEN cDNA 8430419K02 gene |
776 |
0.61 |
| chr2_35623965_35624326 | 0.08 |
Dab2ip |
disabled 2 interacting protein |
1985 |
0.37 |
| chr18_68058494_68058645 | 0.08 |
Gm41764 |
predicted gene, 41764 |
59805 |
0.12 |
| chr6_140888560_140888983 | 0.08 |
Gm30524 |
predicted gene, 30524 |
66838 |
0.11 |
| chr5_96920446_96920776 | 0.08 |
Gm8013 |
predicted gene 8013 |
661 |
0.48 |
| chr7_25613317_25613468 | 0.08 |
Erich4 |
glutamate rich 4 |
2500 |
0.13 |
| chr8_122285085_122285236 | 0.08 |
Zfpm1 |
zinc finger protein, multitype 1 |
3019 |
0.21 |
| chr10_13092588_13092795 | 0.08 |
Plagl1 |
pleiomorphic adenoma gene-like 1 |
1678 |
0.4 |
| chr5_103829138_103829289 | 0.08 |
Aff1 |
AF4/FMR2 family, member 1 |
44854 |
0.11 |
| chr6_113690658_113691420 | 0.08 |
Irak2 |
interleukin-1 receptor-associated kinase 2 |
294 |
0.72 |
| chr16_34056924_34057075 | 0.08 |
Kalrn |
kalirin, RhoGEF kinase |
38988 |
0.16 |
| chr9_111119102_111119253 | 0.08 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
252 |
0.9 |
| chr17_56134469_56135201 | 0.08 |
Sema6b |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
992 |
0.33 |
| chr17_78676930_78677295 | 0.08 |
Strn |
striatin, calmodulin binding protein |
7240 |
0.21 |
| chr6_129183729_129184848 | 0.08 |
Clec2d |
C-type lectin domain family 2, member d |
3673 |
0.14 |
| chr4_19576297_19576523 | 0.08 |
Rmdn1 |
regulator of microtubule dynamics 1 |
1203 |
0.48 |
| chr15_31396603_31396758 | 0.08 |
Gm49300 |
predicted gene, 49300 |
28318 |
0.11 |
| chr15_57793060_57793483 | 0.08 |
9330154K18Rik |
RIKEN cDNA 9330154K18 gene |
54605 |
0.12 |
| chr6_52965535_52965952 | 0.08 |
Gm26215 |
predicted gene, 26215 |
44789 |
0.14 |
| chr10_118839718_118840226 | 0.08 |
Gm47434 |
predicted gene, 47434 |
4866 |
0.15 |
| chr1_133418145_133418838 | 0.08 |
Sox13 |
SRY (sex determining region Y)-box 13 |
5886 |
0.2 |
| chr8_69023381_69023534 | 0.08 |
Gm15716 |
predicted gene 15716 |
21297 |
0.15 |
| chr2_158142922_158143689 | 0.08 |
Tgm2 |
transglutaminase 2, C polypeptide |
3061 |
0.21 |
| chr7_4515813_4516413 | 0.08 |
Tnnt1 |
troponin T1, skeletal, slow |
109 |
0.91 |
| chr2_77169306_77170885 | 0.08 |
Ccdc141 |
coiled-coil domain containing 141 |
481 |
0.83 |
| chr3_30360581_30360732 | 0.08 |
Gm38362 |
predicted gene, 38362 |
86271 |
0.07 |
| chr1_34105041_34105192 | 0.08 |
Mir6896 |
microRNA 6896 |
12243 |
0.17 |
| chr12_83770139_83771096 | 0.08 |
Papln |
papilin, proteoglycan-like sulfated glycoprotein |
6955 |
0.12 |
| chr14_69053224_69053475 | 0.08 |
Gm41192 |
predicted gene, 41192 |
23697 |
0.16 |
| chr14_20929965_20930649 | 0.08 |
Vcl |
vinculin |
909 |
0.62 |
| chr12_111416717_111418003 | 0.08 |
Exoc3l4 |
exocyst complex component 3-like 4 |
70 |
0.96 |
| chr16_85546798_85547372 | 0.08 |
Cyyr1 |
cysteine and tyrosine-rich protein 1 |
3332 |
0.28 |
| chr17_72937316_72937687 | 0.08 |
Lbh |
limb-bud and heart |
16313 |
0.22 |
| chr11_80508926_80509274 | 0.08 |
C030013C21Rik |
RIKEN cDNA C030013C21 gene |
6 |
0.98 |
| chr19_32322600_32322981 | 0.08 |
Sgms1 |
sphingomyelin synthase 1 |
439 |
0.87 |
| chr13_69460904_69461114 | 0.08 |
Gm48676 |
predicted gene, 48676 |
52991 |
0.09 |
| chr7_19163884_19165719 | 0.08 |
Gipr |
gastric inhibitory polypeptide receptor |
293 |
0.77 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.1 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.1 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |